- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2019
Volume: 9, Issue: 12
Cancer Biology
Capillary Nano-immunoassay for Quantification of Proteins from CD138-purified Myeloma Cells
Fluorescence HPLC Analysis of the in-vivo Activity of Glucosylceramide Synthase
Cell Biology
Visualizing Filamentous Actin in Chlamydomonas reinhardtii
Tandem Affinity Purification of SBP-CBP-tagged Type Three Secretion System Effectors
Developmental Biology
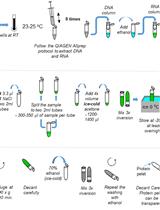
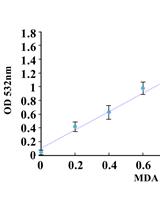
Determining Oxidative Damage by Lipid Peroxidation Assay in Rat Serum
Environmental science
Detachment Procedure of Bacteria from Atmospheric Particles for Flow-cytometry Counting
Microbiology
Production and Purification of Dengue Virus-like Particles from COS-1 Cells
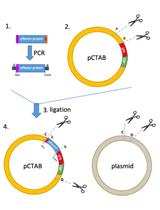
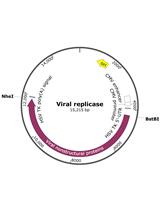
Analysis of Functional Virus-generated PAMP RNAs Using IFNα/β ELISA Assay
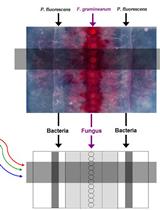
Imaging Gene Expression Dynamics in Pseudomonas fluorescens In5 during Interactions with the Fungus Fusarium graminearum PH-1
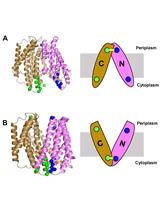
Probing Conformational States of a Target Protein in Escherichia coli Cells by in vivo Cysteine Cross-linking Coupled with Proteolytic Gel Analysis
Molecular Biology
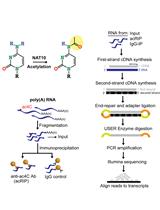
Immunoprecipitation and Sequencing of Acetylated RNA
Neuroscience
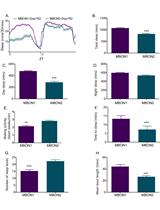
Measurement of Sleep and Arousal in Drosophila
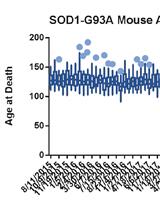
A High-throughput qPCR-based Method to Genotype the SOD1G93A Mouse Model for Relative Copy Number
Rat Model of Empathy for Pain
Live-cell Migration Assays to Study Motility of Neural and Glial (Oligodendrocyte) Progenitor Cells
Optical Stimulation and Electrophysiological Analysis of Regenerating Peripheral Axons
Neurostore: A Novel Cryopreserving Medium for Primary Neurons