- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2019
Volume: 9, Issue: 10
Cancer Biology
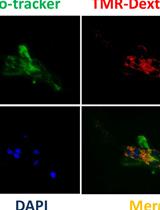
Visualization of Macropinocytosis in Prostate Fibroblasts
Cell Biology
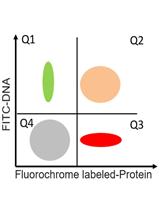
A Flow Cytometric Method to Determine Transfection Efficiency
Measurement of Respiration Rate in Live Caenorhabditis elegans
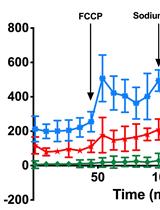
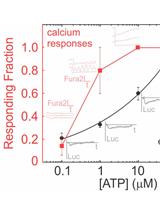
Simultaneous Fluorescent Recordings of Extracellular ATP and Intracellular Calcium in Mammalian Cells
Immunology
TZA, a Sensitive Reporter Cell-based Assay to Accurately and Rapidly Quantify Inducible, Replication-competent Latent HIV-1 from Resting CD4+ T Cells
Hypochlorous Acid Staining with R19-S in the Drosophila Intestine upon Ingestion of Opportunistic Bacteria
Microbiology
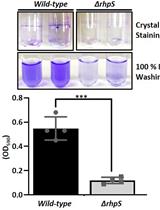
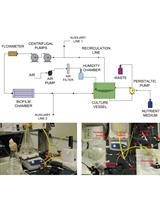
Biofilm Formation Assay in Pseudomonas syringae
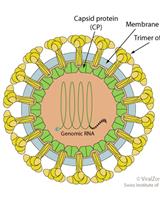
Purification and Proteomic Analysis of Alphavirus Particles from Sindbis Virus Grown in Mammalian and Insect Cells
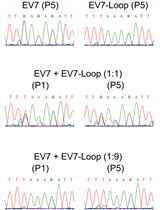
Enterovirus Competition Assay to Assess Replication Fitness
Assembly of a Custom-made Device to Study Spreading Patterns of Pseudomonas putida Biofilms
Neuroscience
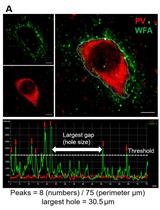
Protocol to Quantitatively Assess the Structural Integrity of Perineuronal Nets ex vivo
Plant Science
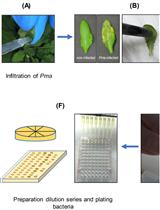
An Improved Bioassay to Study Arabidopsis Induced Systemic Resistance (ISR) Against Bacterial Pathogens and Insect Pests
An Adjustable Protocol to Analyze Chemical Profiles of Non-sterile Rhizosphere Soil