- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2019
Volume: 9, Issue: 3
Biochemistry
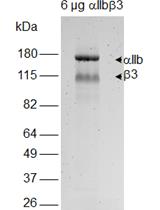
Measurement of Redox States of the β3 Integrin Disulfide Bonds by Mass Spectrometry
Cancer Biology
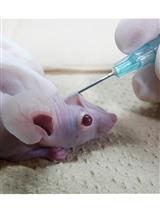
Calvarial Bone Implantation and in vivo Imaging of Tumor Cells in Mice
In situ, Cell-free Protein Expression on Microarrays and Their Use for the Detection of Immune Responses
Cell Biology
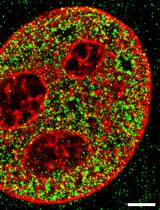
Imaging Higher-order Chromatin Structures in Single Cells Using Stochastic Optical Reconstruction Microscopy
Immunology
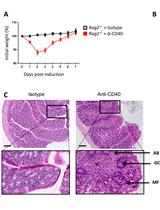
Induction and Analysis of Anti-CD40-induced Colitis in Mice
Microbiology
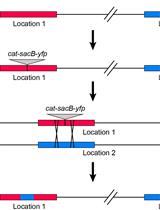
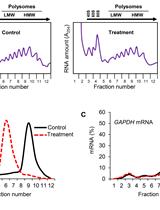
Measuring Homologous Recombination Rates between Chromosomal Locations in Salmonella
Molecular Biology
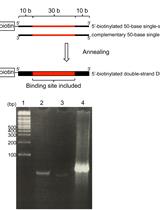
In vitro Protein-DNA Binding Assay (AlphaScreen® Technology)
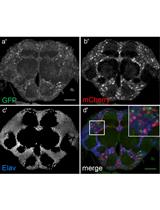
Flip-flop Mediated Conditional Gene Inactivation in Drosophila
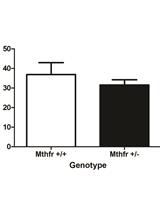
pNP Transgenic RNAi System Manual in Drosophila
Neuroscience
Assessing Spatial Working Memory Using the Spontaneous Alternation Y-maze Test in Aged Male Mice
A Novel Hygrotaxis Assay for Assessing Thirst Perception and Water Sensation in Drosophila
Correction




.jpg)