- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2019
Volume: 9, Issue: 2
Biochemistry
Click Chemistry (CuAAC) and Detection of Tagged de novo Synthesized Proteins in Drosophila
Insulin Tolerance Test under Anaesthesia to Measure Tissue-specific Insulin-stimulated Glucose Disposal
Cell Biology
A Method for Culturing Mouse Whisker Follicles to Study Circadian Rhythms ex vivo
Immunology
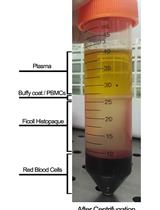
Assessment of Humoral Alloimmunity in Mixed Lymphocyte Reaction
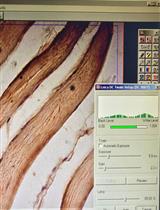
Immunohistochemical Staining of TLR4 in Human Skeletal Muscle Samples
Microbiology
Assessing Yeast Cell Survival Following Hydrogen Peroxide Exposure
Intracellular Invasion and Killing Assay to Investigate the Effects of Binge Alcohol Toxicity in Murine Alveolar Macrophages
Molecular Biology
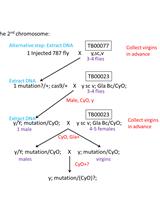
CRISPR-Cas9 Mediated Genome Editing in Drosophila
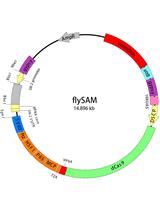
flySAM Transgenic CRISPRa System Manual
Neuroscience
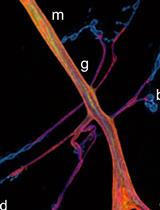
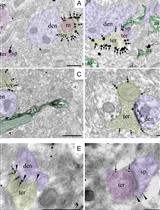
High-resolution Immunoelectron Microscopy Techniques for Revealing Distinct Subcellular Type 1 Cannabinoid Receptor Domains in Brain
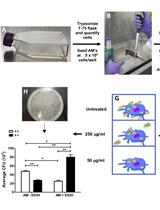
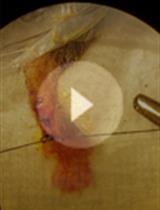
A Mouse Model of Postoperative Pain