- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2017
Volume: 7, Issue: 18
Biochemistry
Lipidomic Analysis of Caenorhabditis elegans Embryos
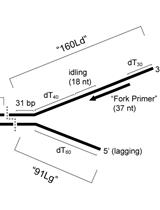
In vitro Assays for Eukaryotic Leading/Lagging Strand DNA Replication
Protease Activity Assay in Fly Intestines
Cancer Biology
Uptake Assays to Monitor Anthracyclines Entry into Mammalian Cells
Cell Biology
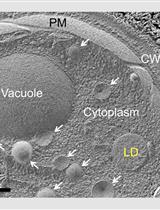
Freeze-fracture-etching Electron Microscopy for Facile Analysis of Yeast Ultrastructure
Immunology
Differentiation of Myeloid-derived Suppressor Cells from Murine Bone Marrow and Their Co-culture with Splenic Dendritic Cells
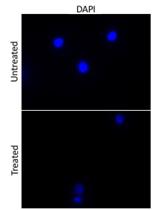
An Assay to Determine Phagocytosis of Apoptotic Cells by Cardiac Macrophages and Cardiac Myofibroblasts
Phagocytosis Assay of Necroptotic Cells by Cardiac Myofibroblasts
Microbiology
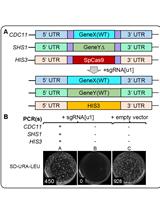
Method for Multiplexing CRISPR/Cas9 in Saccharomyces cerevisiae Using Artificial Target DNA Sequences
Drosophila Fecal Sampling
Neuroscience
Stereotaxic Adeno-associated Virus Injection and Cannula Implantation in the Dorsal Raphe Nucleus of Mice
Preparation of Primary Cultures of Embryonic Rat Hippocampal and Cerebrocortical Neurons
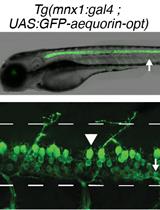
Bioluminescence Monitoring of Neuronal Activity in Freely Moving Zebrafish Larvae
Plant Science
Isolation and Detection of the Chlorophyll Catabolite Hydroxylating Activity from Capsicum annuum Chromoplasts
Detection of Protein S-nitrosothiols (SNOs) in Plant Samples on Diaminofluorescein (DAF) Gels