- Submit a Protocol
- Receive Our Alerts
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Generation of Intestinal Epithelial Monolayers From Single-Cell Dissociated Organoids
Published: Vol 15, Iss 19, Oct 5, 2025 DOI: 10.21769/BioProtoc.5474 Views: 127
Reviewed by: Wendy Leanne HempstockAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols
Monitoring Intestinal Organoid–Derived Monolayer Barrier Functions with Electric Cell–Substrate Impedance Sensing (ECIS)
Sarah Ouahoud [...] Vanesa Muncan
Mar 5, 2024 1323 Views
Primary Neuronal Culture and Transient Transfection
Shun-Cheng Tseng [...] Eric Hwang
Jan 20, 2025 1384 Views
Isolation and Characterization of Cervical Cancer-Associated Mesenchymal Stem Cells From Primary Tumors Using Explant Culture
Surbhi Singla [...] Shalmoli Bhattacharyya
Jun 20, 2025 1817 Views
Abstract
Intestinal organoids are generated from intestinal epithelial stem cells, forming 3D mini-guts that are often used as an in vitro model to evaluate and manipulate the regenerative capacities of intestinal epithelial stem cells. Plating 3D organoids on different substrates transforms organoids into 2D monolayers, which self-organize to form crypt-like regions (which contain stem cells and transit amplifying cells) and villus-like regions (which contain differentiated cells). This “open lumen” organization facilitates multiple biochemical and biomechanical studies that are otherwise complex in 3D organoids, such as drug applications to the cell’s apical side or precise control over substrate protein composition or substrate stiffness. Here, we describe a protocol to generate homogenous intestinal monolayers from single-cell intestinal organoid suspension, resulting in de novo crypt formation. Our protocol results in higher viability of intestinal cells, allowing successful monolayer formation.
Key features
• This protocol requires preexisting experience in culturing mouse intestinal organoids.
• This protocol requires preexisting experience in generating polyacrylamide (PAA) gels for culturing 2D monolayers.
• This protocol generates intestinal monolayers that can be subjected to additional analysis, e.g., drug treatment, immunofluorescent staining, single-molecule fluorescent in-situ hybridization (smFISH), or live imaging.
Keywords: MonolayersBackground
The small intestine is one of the fastest renewing tissues in the body. Intestinal epithelial cells cover recurring units of crypts- invaginations into the intestinal stroma and villi finger-like protrusions that extend into the gut lumen. The crypts contain intestinal stem cells, Paneth cells, and transit amplifying cells. As cells exit the crypt, they differentiate into absorptive or secretory cell types. Intestinal epithelial cells adhere and migrate on the basement membrane, a sheet-like extracellular matrix. Both biochemical and mechanical cues, often originating in the extracellular matrix or surrounding cell types, maintain the behavior of the different cell populations along the crypt–villus axis during homeostasis [1,2].
The use of intestinal organoids emerged over a decade ago, as it was demonstrated that intestinal stem cells can form mini-guts–intestinal organoids, which recapitulate the stem cell–containing crypt-like compartment as well as regions of differentiated cells, similarly to cells covering the villi [3]. These organoids can be expanded indefinitely in 3D culture due to the self-renewing capacity of intestinal stem cells. While intestinal organoids are a powerful tool to investigate stem cell biology, their 3D closed lumen structure limits access to the cell’s apical surface, hinders solute transport studies, and makes it difficult to precisely control the extracellular environment, such as stiffness or protein composition.
To address these limitations, several methods have been developed to convert 3D organoids into 2D intestinal monolayers [4–7]. These use intestinal organoids as a starting material; when plated on a 2D structure, intestinal organoids will form a self-organizing 2D monolayer with crypt-like regions that contain stem cells surrounded by differentiated cells, mimicking the crypt–villus axis.
These monolayers allow easy drug or nutrient application, live imaging, and techniques such as transepithelial electric resistance (TEER) or electric cell-substrate impedance sensing (ECIS) [8–10]. Additionally, they enable controlled studies of mechanical cues by varying substrate stiffness or coating [11,12], for example, by using polyacrylamide (PAA) gels, and allow measurement of cellular forces via traction force microscopy [4].
Intestinal monolayers can be generated by plating small clumps of dissociated organoids. Alternatively, organoids can be dissociated into single cells, which then form monolayers through de novo crypt formation [4]. Here, we describe a robust protocol using Accumax or Accutase to dissociate organoids into single cells. This method improves cell viability and enhances the attachment efficiency of seeded cells. It enables the formation of uniform, self-organizing intestinal monolayers from either single or mixed cell populations. These monolayers can be used for various downstream applications, including drug treatments, live imaging, immunofluorescence, and smFISH. The same protocol also supports simple adhesion assays to assess cell-substrate interactions on different coatings.
Materials and reagents
Biological materials
1. Mouse intestinal organoids [13], grown in 50 μL Matrigel drops in a 24-well plate; see General note 1
Reagents
1. DPBS (Thermo Fisher scientific, catalog number: 14190136)
2. Gibco DMEM F12-Glutamax (Thermo Fisher scientific, catalog number: 11514436)
3. Gibco 100× antibiotic-antimycotic (Thermo Fisher scientific, catalog number: 15240096)
4. B27 supplement 50×, serum-free (Thermo Fisher scientific, catalog number: 17504001)
5. N2 supplement 100× (Thermo Fisher scientific, catalog number: 15410294)
6. Mouse EGF (Peprotech, catalog number: 17872513)
7. Mouse FGF (Peprotech, catalog number: 17844573)
8. Murine Noggin (Peprotech, catalog number: 250-38)
9. R-spondin (R&D Systems, catalog number: 3474-RS-050)
10. Nicotinamide (Sigma-Aldrich, CAS: 98-92-0)
11. Chir-99021 (Euromedex, catalog number: AB-M1692-100MG; CAS: 252917-06-9)
12. Rock inhibitor Y27632 (ATCC, catalog number: ATCC-ACS-3030)
13. Accumax (Sigma-Aldrich, catalog number: A7089)
14. Trypan blue stain (Invitrogen, catalog number: T10282)
15. Collagen I, rat tail, 100 mg (Corning, catalog number: 354236)
Solutions
1. Stock solutions
2. Cell dissociation solution (see Recipes)
3. Stop solution (see Recipes)
4. ENR medium (see Recipes)
5. Plating medium (see Recipes)
6. ENR-CNY medium (see Recipes)
Recipes
1. Stock solutions
| Reagent | Stock concentration | Solvent |
|---|---|---|
| Chir-99021 | 3 mM | DMSO |
| Y27632 | 1 mM | Distilled water |
| Nicotinamide | 1 M | Distilled water |
| EGF | 200 μg/mL | PBS |
| FGF | 100 μg/mL | PBS |
Aliquot and store at -20 °C.
2. Cell dissociation solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| Accumax | - | 1 mL |
| Chir-99021 (3 mM stock) | 3 μM | 1 μL |
| Y27632 (1 mM stock) | 10 μM | 1 μL |
For 12 Matrigel drops, 1 mL of dissociation solution is needed. Keep dissociation solution on ice. Make fresh each time.
3. Stop solution
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| DMEM F12-Glutamax | - | 1 mL |
| B27 50× | 1× | 20 μL |
| Chir-99021 (3 mM stock) | 3 μM | 1 μL |
| Y27632 (1 mM stock) | 10 μM | 1 μL |
For every 1 volume of dissociation solution, 2 volumes of stop solution are needed. For 1 mL of dissociation solution, 2 mL of stop solution is needed. Pre-heat the stop solution to 37 °C. Make fresh each time.
4. ENR medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| DMEM F12-Glutamax | - | - |
| Antibiotic-antimycotic | 2% | - |
| Mouse EGF | 10 ng/μL | - |
| Noggin | 100 ng/μL | - |
| R-spondin | 500 ng/μL | - |
| FGF | 10 ng/μL | - |
| N2 100× | 1× | - |
| B27 50× | 1× | - |
ENR medium can be kept at 4 °C for 2–3 weeks and will be used as the base medium for the monolayers throughout the experiment. Use 700 μL for each fluorodish.
5. Plating medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| ENR medium | - | - |
| Chir-99021 (3 mM stock) | 3 μM | - |
| Y27632 (1 mM stock) | 10 μM | - |
| - | - | 45–50 μL for each plate |
ENR medium can be kept at 4 °C for 2–3 weeks. Each gel should be plated with 45 μL of plating medium + cells. Add Chir and Y27 to the ENR medium at the needed volume right before plating.
6. ENR-CNY medium
| Reagent | Final concentration | Quantity or volume |
|---|---|---|
| ENR medium | - | - |
| Chir-99021 (3 mM stock) | 3 μM | - |
| Y27632 (1 mM stock) | 10 μM | - |
| Nicotinamide (1 M stock) | 10 mM | - |
| - | - | 700 μL for each plate |
Use this medium for cell spreading in the first 48–72 h. Use 700 μL for each fluorodish. Pre-heat to 37 °C. ENR medium can be kept at 4 °C for 2–3 weeks. Add Chir, Y27, and Nicotinamide to the ENR medium at the needed volume right before adding medium to the plated cells.
Laboratory supplies
1. 35 mm glass‐bottom dish (WPI FluoroDish FD35‐100) with 18 mm polyacrylamide gels (PAA), coated with collagen type-1 and washed with PBS, or any alternative substrate for cell attachment. See Baghdadi et al. [14], supplementary table. See General notes 2 and 3.
2. 20 G needle (TERUMO, catalog number: AN*2038R1)
3. 23 G needle (BD Microlance, catalog number: 300800)
4. 24 G needle (TERUMO, catalog number: NN-2425R)
5. 10 mL syringe (TERUMO, catalog number: MDSS10SE)
6. 2.5 mL syringe (TERUMO, catalog number: SS*02SE1)
7. Cell strainer 40 μm (FisherBrand, catalog number: 22363547)
8. 15 mL conical tubes (Sarstedt, catalog number: 62.554.002)
9. 50 mL conical tubes (Sarstedt, catalog number: 62.548.101)
10. 10 μL pipette tips (STARLAB, catalog number: S1121-2710)
11. 200 μL pipette tips (STARLAB, catalog number: S1120-8710)
12. 1,000 μL pipette tips (STARLAB, catalog number: S1122-1730)
Equipment
1. Laboratory centrifuge with rotors for 15 mL conical tubes, temperature-controlled, chilled to 4 °C (Eppendorf Centrifuge, model: 5804 R)
2. Cell incubator
3. Cell counter (DeNovix CellDrop FL counter)
4. Microscope for evaluating cell dissociation
Procedure
A. Organoid dissociation
1. For each gel, you will need 4–6 Matrigel drops with mature organoids (drop volume is 50 μL, containing mature organoids, cultured for 2–3 days). Aspirate growth medium from around the Matrigel drop. Add 500 μL of cold DPBS.
2. Using a 1 mL pipette tip, a cell scraper, or by pipetting the DPBS up and down, detach the Matrigel drop and the organoids inside it from the plate. Collect the organoids, broken Matrigel, and DPBS into a 50 mL Falcon.
3. Using a 20 G needle and a 10 mL syringe, push the solution of organoids up and down 10 times to assist the release of the organoids from the Matrigel and ease their dissociation.
4. Spin at 300× g for 5 min at 4 °C. Discard the supernatant.
5. Resuspend the pellet with cell dissociation solution. Use 1 mL of dissociation solution for every 12 Matrigel drops used. Transfer the organoid solution to a 5 or 15 mL tube.
6. Place the organoid solution at 37 °C for 10–13 min. Every 4–5 min, flick the tube and pipette the solution up and down to help the dissociation process.
7. For separating cell clumps into single cells, pass the cell solution through a 23 G needle and a 2.5 mL syringe 10 times. Change the needle to a 24 G and pass the solution through the syringe an additional 5–10 times.
8. Monitor the dissociated cells under the microscope; if cells remain clumped, pass the solution several more times through the 24 G needle, expelling the cells through the needle, while the needle is pressed against the side of the tube.
9. Once cell dissociation is complete, stop the reaction by adding 1 volume of stop solution (if you have used 1 mL of dissociation solution, use 1 mL of stop solution now).
10. Filter the cells using a 40 μm filter to reach a single-cell suspension. Note that filtering will reduce the final number of cells seeded, as some will be lost on the filter (see General note 4).
11. Spin the cells at 400× g for 5 min at 4 °C.
12. Discard the supernatant, resuspend the pellet in 1 volume of fresh stop solution, and spin the cells again. Optional: Use a lower G (170) to ease cell separation before plating.
Optional: Take a few microliters to measure cell numbers and viability, using trypan blue stain.
Optional: Dissociated cells can be used for different applications; see General note 5.
B. Cell plating
1. Wash the PAA gel with fresh PBS (at room temperature); aspirate the PBS, making sure not to damage the gel. Place the gel to dry lightly in the hood during the last cell centrifugation (step A12) for approximately 5 min. Do not over-dry the gels. If using a different cell substrate, lightly dry it before plating to allow for placing a small concentrated drop of cells on top of the substrate. See General note 6 and Figure 1A.
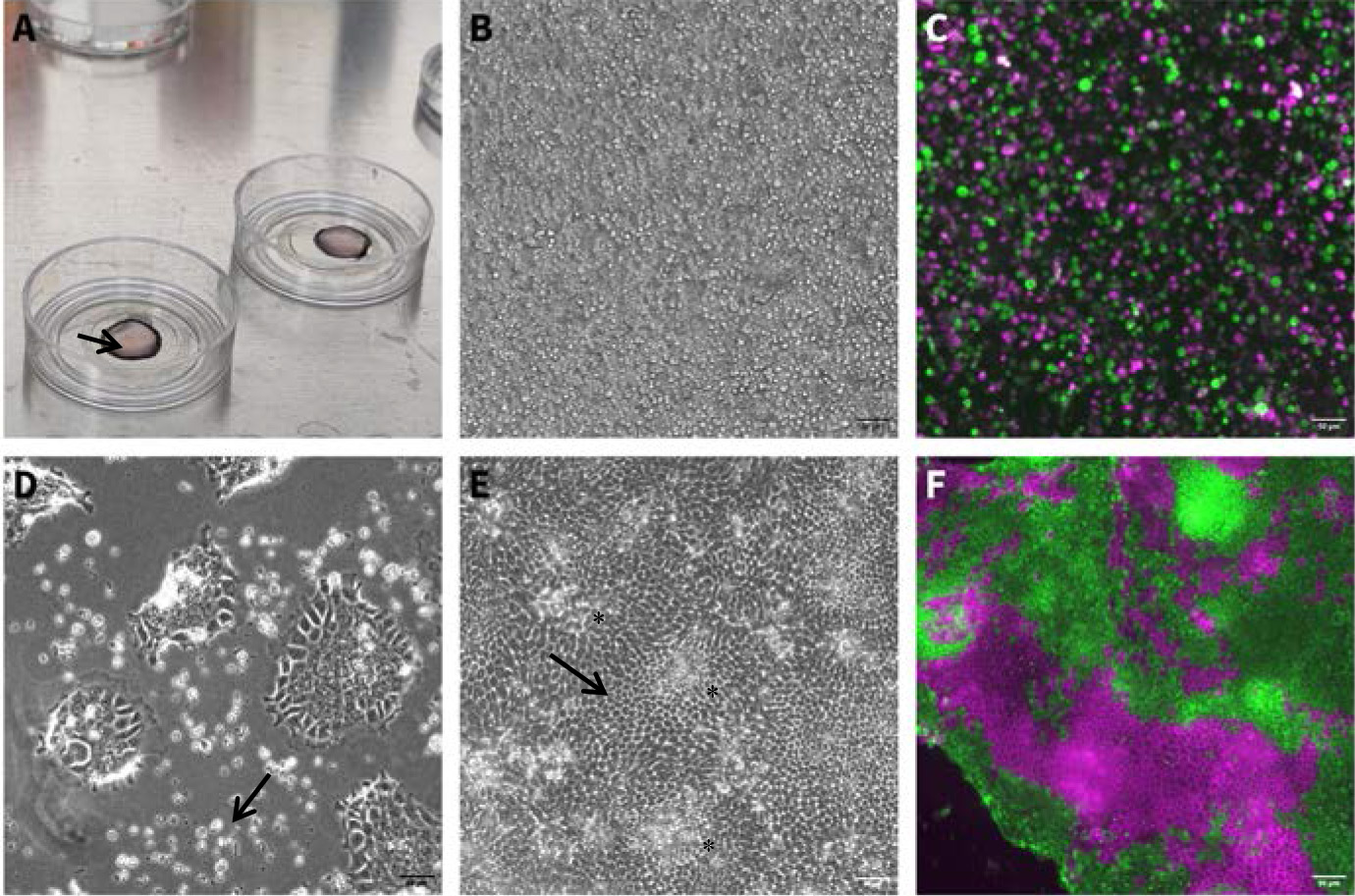
Figure 1. Culture morphology during single-cell monolayer formation. (A) Medium containing single-cell suspension on a PAA gel (gel appears as a circle, arrow pointing to the gel rim), after plating (step B2). (B) Brightfield image of single-cell suspension after initial incubation time (step B3). Scale bar: 50 μm. (C) Fluorescent image of single-cell suspension generated from a mix of two different organoid lines at the end of the initial incubation time (step B3). Scale bar: 50 μm. (D) Cell attachment following 48 h in ENR-CNY medium (step B6). Dead cells appear as white, round clumps (arrow). Attached cells are flat and mesenchymal-looking due to the use of the plating medium, which facilitates cell spreading. Scale bar: 50 μm. (E) Brightfield image of mature monolayer, 10 days after initial seeding. Some crypt regions are marked with an asterisk. Note the hexagonal shape in the villus-like regions (arrow). Scale bar: 50 μm. (F) Fluorescent image of a mature monolayer from mixed organoids (seen in C1), expressing tdTomato or GFP. Scale bar: 50 μm.
2. Resuspend the cell pellet from step A12 in plating medium. For each gel, plate approximately 500,000–800,000 cells in 45–50 μL of plating medium. See General note 7 and Figure 1A.
3. Place the gels with the drop of cells in a cell-culture incubator at 37 °C with 5% CO2 for 1–2 h, allowing the cells to attach.
4. Pre-heat ENR-CNY medium to 37 °C. Prepare 700 μL for each plate.
5. When the initial incubation is over and cells have had time to attach, add the pre-heated ENR-CNY medium to the plate, disturbing the drop of cells as little as possible but making sure the gel and plate are completely covered with medium. ENR-CNY medium allows the cells to spread and form a homogenous monolayer. See Figure 1B, C.
6. Twenty-four to seventy-two hours after plating, some of the cells should have properly attached and formed a monolayer. Dead cells will appear as dots on the gel or will float in the medium. Gently tilt the plate and aspirate the ENR-CNY medium using a pipette and replace it with pre-heated ENR medium to allow the cells to mature and differentiate. Make sure not to disturb the cells. See Figure 1D.
7. Monitor monolayer growth and differentiation, changing media gently, using a pipette to aspirate old medium and supplying new medium every 1–2 days. See General notes 8 and 9. See Figure 1E, F for a mature and differentiated monolayer image.
Validation of protocol
Monolayers can be examined under the microscope for maturation and differentiation. In crypt-like regions, cells will appear dense and arrange in a circular pattern (Figure 2A), while differentiated cells will assume a cuboid or hexagonal shape (Figure 1E). In addition, monolayers can be stained with different antibodies to identify cell types, as well as to validate the formation of crypt-like regions and differentiated cells. For a list of possible antibodies, see Table 1 and Figure 2.
Table 1. Recommended antibodies
| Cells in crypt region | Differentiated cells |
|---|---|
| Paneth cells, Sox9 (1:100, #AF3075, Bio-Techne) | AldoB (1:200, #ab75751, Abcam) |
| Stem cells, Olfm4 (1:150, #39141, Cell Signalling) | Cytokeratin (1:100) (#Z0622, Dako) |
| Paneth cells, Lyzozyme (1:1,000, #A0099, Dako) |
Additionally, as cell proliferation in mature monolayers occurs only in the crypt-like region, one could pulse-chase cell proliferation using EDU or stain with KI67 antibodies. Monolayers can be fixed using 4% PFA/PBS for 30 min at room temperature, though different fixation times/reagents might be needed for different antibodies.
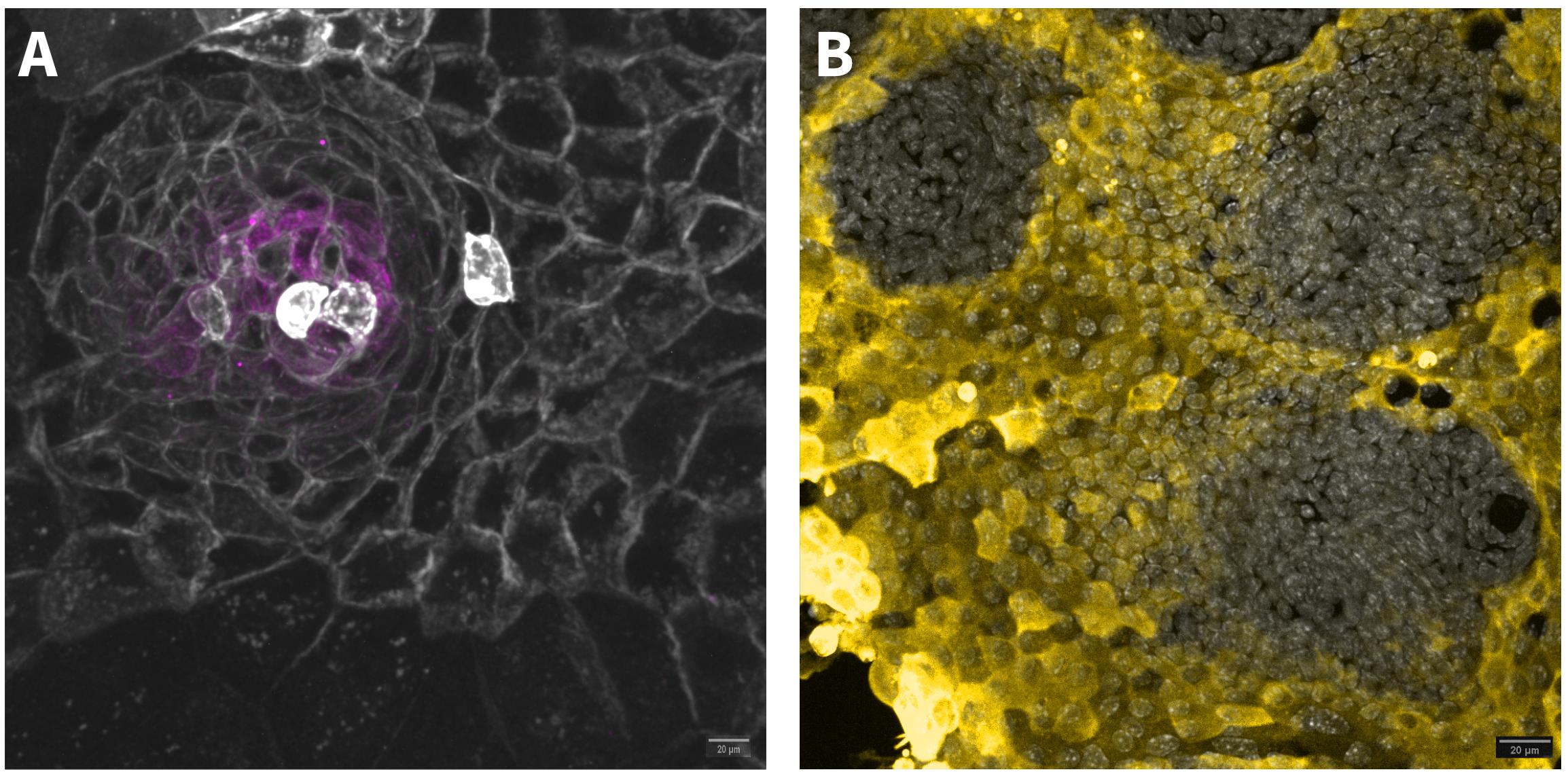
Figure 2. Antibody staining to validate crypt-like region and differentiated cells. (A) OLFM4 (magenta) staining in a crypt-like region. The monolayer was made from non-induced, tdTomato-expressing mT/mG (cre-recombinase-activated membrane-tomato/membrane GFP) organoids (grey). Scale bar: 20 μm. (B) Differentiated cells stained with AldoB antibody (yellow). Nuclei are stained with DAPI (grey). Scale bar: 20 μm.
This protocol has been used and validated in the following research article(s):
• Baghdadi et al. [14]. PIEZO-dependent mechanosensing is essential for intestinal stem cell fate decision and maintenance. Science (Figure 3H).
General notes and troubleshooting
General notes
1. Organoids can be grown as hyperproliferative cysts, thereby increasing the number of cells with the potential of forming de novo crypts. To that end, newly split organoids should be grown in ENR containing 10 μM Chir990201 and 10 mM Nicotinamide. Hyperproliferative crypts have a round, cystic morphology.
2. This protocol uses PAA gels as a substrate for cell growth; however, other matrices can also be used, such as agarose gels, collagen gels, or Matrigel.
3. Generation of PAA gels and protein coating, as well as an alternative method to generate monolayers from single-cell suspension, is described in Pérez-González et al. [4]. Coating of the PAA gels with Collagen type-1 is necessary for cell attachment on PAA gels and cannot be omitted. We recommend using 250–500 μg/mL coat overnight at 4 °C. Note that gel stiffness, as well as additional protein coats, can affect the efficiency of cell attachment; this protocol has been tested successfully with 2 and 5 kPA PAA gels, with gels coated, in addition to Collagen1, with Laminin111.
4. The filtering stage can be skipped, and cells can be plated after the second wash. In this case, some clumped cells will remain, but cell numbers will be higher.
5. Dissociated cells can be used for other applications other than forming monolayers, such as FACS sorting or cell adhesion assays.
6. Make sure the gels are not too dry, and that the drop does not exceed the size of the gel. If the drop is not sustained on the gel but spreads, it means the gel was not dry enough. Try to aspirate the cells, wash the gel with PBS, dry the gel again, and reapply the cell drop.
7. Cells can be resuspended in higher volumes (>50 μL per gel), though a crowded-cell drop is beneficial for cell attachment.
8. Media change should be performed delicately. Tilt the plate and aspirate old media using a pipette; wash gently, releasing the media one drop at a time on top of the monolayer to gently release dead cells. Repeat this 3–4 times. Then, aspirate and discard the old media and add new media without releasing the media directly on the cells.
9. Mature, differentiated monolayers can be kept in culture for over 10 days. Pre-patterning of the collagen on top of the gel or using a smaller substrate area for cell plating to limit monolayer boundaries is beneficial for keeping the monolayer for long periods of time and for achieving a more homogenous monolayer.
Troubleshooting
Problem 1: Few cells after cell dissociation.
Possible cause A: Poor collection of organoids from the Matrigel drop.
Solution A: Make sure to properly scrape the Matrigel drop from the plate, as one would do during organoid splitting. Check the plate under the microscope to make sure no organoids were left stuck to the plate. Make sure not to aspirate the pellet after centrifugation.
Possible cause B: Cells were not broken down into single cells before filtering and were stuck on the filter.
Solution B: Repeat cell dissociation with a 24 G needle until the cell suspension is broken into single cells. Monitor cell dissociation under a microscope before proceeding.
Problem 3: Cell clumps seen in cell drop during plating.
Possible cause: During the last centrifugation, cells clumped together.
Solution: Resuspend the pellet thoroughly by pipetting up and down in the medium before plating. If necessary, resuspend using a 24 G needle. Another possibility is to centrifuge; during the second centrifugation, use a lower G to minimize cells sticking together.
Problem 4: No cell attachment after incubation.
Possible cause: Poor collagen coating, poor preparation of substrate used for plating.
Solution: Make sure to coat the PAA gels with Collagen type 1. We recommend using 250–500 μg/mL coat overnight at 4 °C. Alternatively, modify the cell substrate.
Problem 5: No cell attachment/no monolayer formation.
Possible cause: Low cell density during plating/poor viability after dissociation.
Solution: Use more Matrigel drops per gel or more organoids for making the cell suspension. Another possibility is to grow organoids as cysts to increase the number of stem cells that can form the monolayer. Work faster during the dissociation phase and omit filtering of the cell suspension before plating. Do not increase plating volume above 45 μL per gel. Additionally, adding DNase to the dissociation media can improve cell viability, although this is not mandatory.
Acknowledgments
The work on this protocol was funded by the European Union’s Horizon Europe research and innovation program under the Marie Skłodowska-Curie grant agreement No. 101066253 and by the Agence Nationale de la Recherche (ANR-23-CE14-0030).
The establishment of this protocol was assisted by the work presented in Pérez-González et al. [4] and Jacquemin et al. [15]. This protocol was used in Baghdadi et al. [14].
Competing interests
The authors declare that they have no competing interests.
References
- Barker, N. (2014). Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 15(1): 19–33. https://doi.org/10.1038/nrm3721
- Simon-Assmann, P., Kedinger, M., De Arcangelis, A., Rousseau, V. and Simo, P. (1995). Extracellular matrix components in intestinal development. Experientia. 51(9–10): 883–900. https://doi.org/10.1007/BF01921739
- Sato, T., Stange, D. E., Ferrante, M., Vries, R. G. J., van Es, J. H., van den Brink, S., van Houdt, W. J., Pronk, A., van Gorp, J., Siersema, P. D., et al. (2011). Long-term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology. 141(5): 1762–1772. https://doi.org/10.1053/j.gastro.2011.07.050
- Pérez-González, C., Ceada, G., Greco, F., Matejčić, M., Gómez-González, M., Castro, N., Menendez, A., Kale, S., Krndija, D., Clark, A. G., et al. (2021). Mechanical compartmentalization of the intestinal organoid enables crypt folding and collective cell migration. Nat Cell Biol. 23(7): 745–757. https://doi.org/10.1038/s41556-021-00699-6
- Altay, G., Larrañaga, E., Tosi, S., Barriga, F. M., Batlle, E., Fernández-Majada, V. and Martínez, E. (2019). Self-organized intestinal epithelial monolayers in crypt and villus-like domains show effective barrier function. Sci Rep. 9(1): 10140. https://doi.org/10.1038/s41598-019-46497-x
- Sanman, L. E., Chen, I. W., Bieber, J. M., Steri, V., Trentesaux, C., Hann, B., Klein, O. D., Wu, L. F. and Altschuler, S. J. (2021). Transit-Amplifying Cells Coordinate Changes in Intestinal Epithelial Cell-Type Composition. Dev Cell. 56(3): 356–365.e9. https://doi.org/10.1016/j.devcel.2020.12.020
- Thorne, C. A., Chen, I. W., Sanman, L. E., Cobb, M. H., Wu, L. F. and Correspondence, S. J. A. (2018). Enteroid Monolayers Reveal an Autonomous WNT and BMP Circuit Controlling Intestinal Epithelial Growth and Organization. Dev Cell. 44: 624–633.e4. https://doi.org/10.1016/j.devcel.2018.01.024
- Wegener, J., Keese, C. R. and Giaever, I. (2000). Electric Cell–Substrate Impedance Sensing (ECIS) as a Noninvasive Means to Monitor the Kinetics of Cell Spreading to Artificial Surfaces. Exp Cell Res. 259(1): 158–166. https://doi.org/10.1006/excr.2000.4919
- Anwer, S. and Szászi, K. (2020). Measuring Cell Growth and Junction Development in Epithelial Cells Using Electric Cell-Substrate Impedance Sensing (ECIS). Bio-Protoc. 10(16): e3729. https://doi.org/10.21769/BioProtoc.3729
- Srinivasan, B., Kolli, A. R., Esch, M. B., Abaci, H. E., Shuler, M. L. and Hickman, J. J. (2015). TEER Measurement Techniques for In Vitro Barrier Model Systems. SLAS Technol. 20(2): 107–126. https://doi.org/10.1177/2211068214561025
- He, S., Lei, P., Kang, W., Cheung, P., Xu, T., Mana, M., Park, C. Y., Wang, H., Imada, S., Russell, J. O., et al. (2023). Stiffness Restricts the Stemness of the Intestinal Stem Cells and Skews Their Differentiation Toward Goblet Cells. Gastroenterology. 164(7): 1137–1151.e15. https://doi.org/10.1053/j.gastro.2023.02.030
- Ramadan, R., Wouters, V. M., Van Neerven, S. M., De Groot, N. E., Garcia, T. M., Muncan, V., Franklin, O. D., Battle, M., Carlson, K. S., Leach, J., et al. (2022). The extracellular matrix controls stem cell specification and crypt morphology in the developing and adult mouse gut. Biol Open. 11(12): bio059544. https://doi.org/10.1242/bio.059544
- O’Rourke, K. P., Ackerman, S., Dow, L. E. and Lowe, S. W. (2016). Isolation, Culture, and Maintenance of Mouse Intestinal Stem Cells. Bio-Protoc. 6(4): e1733. https://doi.org/10.21769/bioprotoc.1733
- Baghdadi, M. B., Houtekamer, R. M., Perrin, L., Rao-Bhatia, A., Whelen, M., Decker, L., Bergert, M., Pérez-Gonzàlez, C., Bouras, R., Gropplero, G., et al. (2024). PIEZO-dependent mechanosensing is essential for intestinal stem cell fate decision and maintenance. Science. 386(6725): eadj7615. https://doi.org/10.1126/science.adj7615
- Jacquemin, G., Wurmser, A., Huyghe, M., Sun, W., Homayed, Z., Merle, C., Perkins, M., Qasrawi, F., Richon, S., Dingli, F., et al. (2022). Paracrine signalling between intestinal epithelial and tumour cells induces a regenerative programme. eLife. 11: e76541. https://doi.org/10.7554/eLife.76541
Article Information
Publication history
Received: Jul 8, 2025
Accepted: Sep 2, 2025
Available online: Sep 19, 2025
Published: Oct 5, 2025
Copyright
© 2025 The Author(s); This is an open access article under the CC BY-NC license (https://creativecommons.org/licenses/by-nc/4.0/).
How to cite
Felsenthal, N. and Vignjevic, D. M. (2025). Generation of Intestinal Epithelial Monolayers From Single-Cell Dissociated Organoids. Bio-protocol 15(19): e5474. DOI: 10.21769/BioProtoc.5474.
Category
Stem Cell > Organoid culture
Cell Biology > Cell isolation and culture > Monolayer culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Tips for asking effective questions
+ Description
Write a detailed description. Include all information that will help others answer your question including experimental processes, conditions, and relevant images.
Share
Bluesky
X
Copy link








