- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Shikimate Hydroxycinnamoyl Transferase (HCT) Activity Assays in Populus nigra
Published: Vol 3, Iss 22, Nov 20, 2013 DOI: 10.21769/BioProtoc.978 Views: 14881
Reviewed by: Tie Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1474 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3209 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1824 Views
Abstract
Lignin is a complex phenolic polymer deposited in secondarily-thickened plant cell walls. The polymer is mainly derived from the three primary monolignols: p-coumaryl, coniferyl and sinapyl alcohol which give rise to p-hydroxyphenyl, guaiacyl and syringyl units (H, G and S units, respectively) when coupled into the polymer. The building blocks differ in their degree of methoxylation and their biosynthetic pathway is catalyzed by more than 10 enzymes. HCT plays a crucial role by channeling the phenylpropanoids towards the production of coniferyl and sinapyl alcohols. Interestingly, HCT has been reported to be implicated in the pathway both upstream and downstream of the 3-hydroxylation of the aromatic ring of p-coumaroyl shikimate (Figure 1) (Hoffmann et al., 2003; Hoffmann et al., 2004; Vanholme et al., 2013b). These features highlight the importance of developing an assay to reliably measure HCT activity in planta. Here, we describe a UPLC-MS-based method for the analysis of HCT activity in xylem total protein extracts of Populus nigra, which can be adapted to other woody and herbaceous plant species. The protocol was initially described in Vanholme et al. (2013a).
Keywords: enzyme activity 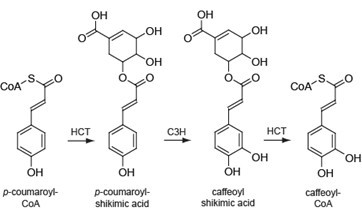
Figure 1. The two enzymatic reactions of the phenylpropanoid pathway catalyzed by HCT. HCT (AEN02914) converts p-coumaroyl-CoA into p-coumaroyl shikimic acid (first HCT-reaction), which is converted to caffeoyl shikimic acid by C3H (coumarate 3-hydroxylase). HCT converts it further to caffeoyl-CoA (second HCT-reaction).
Materials and Reagents
- Tris base (2-amino-2-hydroxymethyl-propane-1,3-diol) (Biosolve, catalog number: 77-86-1 )
- Dithiothreitol (DTT) (AG Scientific, catalog number: C-1029 )
- Polyvinylpolypyrrolidone (PVPP) (Sigma-Aldrich, catalog number: P-6755 )
- Glycerol (Sigma-Aldrich, catalog number: G-7893 )
- Complete Mini Protease Inhibitor Cocktail Tablets (Roche, catalog number: 0 4693159001 )
- Bio-Rad Protein Assay (Bio-Rad Laboratories, catalog number: 500-0006 )
- Bovine Serum Albumin (BSA) (Sigma-Aldrich, catalog number: A-7906 )
- p-coumaroyl-CoA (TransMIT, catalog number: C030 )
- Shikimic acid (Sigma-Aldrich, catalog number: S-5375 )
- Caffeoyl shikimate (AnalytiCon Discovery GmbH, catalog number: NP-00058 )
- Coenzyme-A hydrate (CoA) (Sigma-Aldrich, catalog number: A-3164 )
- Acetonitrile ULC/MS grade (Biosolve, catalog number: BIO-012041 )
- Formic acid ULC/MS grade (Biosolve, catalog number: BIO-06914131 )
- Liquid chromatography (LC) sample vials (Waters, catalog numbers: 186002639 and 2639531020 )
- Ice
- Liquid nitrogen
- Protein Extraction Buffer (see Recipes)
- Reaction Mix (see Recipes)
Equipment
- Nunc 96-well microplate without lid and flat bottom wells (Thermo Fisher Scientific, catalog number: 269787 )
- Safe-Lock tubes 2.0 ml (Eppendorf, catalog number: 3706 )
- Stainless steel surgical scalpel blades No.22 (Swann-Morton, catalog number: 0 308 )
- Mortar (150 x 70 mm; 700 ml) (The Morgan Advanced Materials Company plc, catalog number: 12906-6a )
- Pestle (150 x 36 mm) (Haldenwanger, catalog number: 12906-2 )
- Balance (Mettler Toledo, model: XP-105 Delta Range )
- Vortex (IKA, model: MS2 Minishaker L002050 )
- Temperature controlled benchtop microcentrifuge (Eppendorf, model: 5417R )
- Temperature controlled microplate spectrophotometer (Molecular Devices, model: Spectra Max 250 )
- Thermoblock (Eppendorf Thermomixer Compact) or water bath
- UPLC-MS system (In our case: Waters Acquity UPLC system (WATERS) connected to a Thermo LTQ XL mass spectrometer (Thermo Fisher Scientific) or a Synapt HDMS Q-Tof (WATERS). Chromatographic separation was performed on a Waters Acquity BEH RP C18 (2.1 mm x 150 mm, 1.7 μm) column (WATERS))
- Ultrafreezer
- Optional: Qubit 2.0 Fluorometer (Invitrogen)
Software
- XCalibur 2.0 software (Thermo Fisher Scientific, Waltham) was used to acquire, analyze and manage mass spectrometry information
Procedure
- Protein extraction
- Collect 15-cm stem segments derived from the base of 1-m tall sprouts emerging from the trunk of a poplar tree using secateurs. If 1-year old trees are used, segments of the main stem can be used.
- Immediately submerge the fresh plant tissue into liquid nitrogen and store the samples at -80 °C until further use.
- Incubate the frozen stem segments on ice for a few minutes and make a small incision in the bark on one end of the segment. The bark can be easily pealed from the underlying tissue. Some part of the cambium can stick to the debarked stem, but can be easily removed by gently scrubbing the stem with a scalpel. Scrape the xylem tissue using a scalpel, making sure the scraped tissue is immediately collected in a precooled mortar (use liquid nitrogen to keep the material frozen during sampling).
- Grind the scraped xylem tissue to a fine powder using a mortar and pestle.
- For xylem total protein extraction, weigh ~100 mg of ground xylem tissue in a precooled 2-ml tube and add 1 ml of ice-cold protein extraction buffer. The composition of the protein extraction buffer is given as Recipe 1, at the end of the protocol.
- Vortex thoroughly and incubate on ice for 1 h, inverting the tubes every 5-10 min to prevent precipitation.
- Centrifuge at 20,000 x g for 10 min at 4 °C and transfer the supernatant to a new precooled tube. Keep all samples on ice.
- Collect 15-cm stem segments derived from the base of 1-m tall sprouts emerging from the trunk of a poplar tree using secateurs. If 1-year old trees are used, segments of the main stem can be used.
- Protein quantification using the Bradford method (Bradford, 1976)
- Prepare the BSA standards for the calibration curve as follows:
Volume of BSA (100 μg/ml)
Volume of Water
BSA Concentration
0 μl
240 μl
0 μg/ml
6 μl
234 μl
2.5 μg/ml
12 μl
228 μl
5 μg/ml
18 μl
222 μl
7.5 μg/ml
24 μl
216 μl
10 μg/ml
27 μl
213 μl
11.25 μg/ml
30 μl
210 μl
12.5 μg/ml
- For the calibration curve, add 240 μl of each standard (above) to a separate well of a 96-well flat bottom microplate. All measurements are performed in triplicate.
- For the quantification of xylem total protein samples, add 210 μl of water in each well and 30 μl of the protein extract dilutions in triplicate. Prepare the dilutions using the protein extraction buffer, ranging from 5x to 100x diluted (depending on the amount of plant material used for the extraction).
- Add 60 μl of Bio-Rad Protein Assay reagent to each well and incubate at room temperature for 5 min.
- Read absorbance at 595 nm (A595) on a plate spectrophotometer.
For the calculation of protein concentrations, generate a standard curve by plotting the BSA concentration (X-axis) versus A595 (Y-axis). After obtaining the trend line, use its corresponding equation and the absorbance of the protein sample to resolve the unknown concentration. The correlation coefficient of the trend line should be close to 1.00 (preferentially above 0.97) and all measured absorbances of the protein samples should fall into the linear range.
Notes:
- Figure 2 shows an example of protein quantification using the Bradford method.
- As alternative to Bradford, Qubit (Invitrogen) can be used according manufacturer’s instructions for accurate and efficient quantification of protein concentrations.
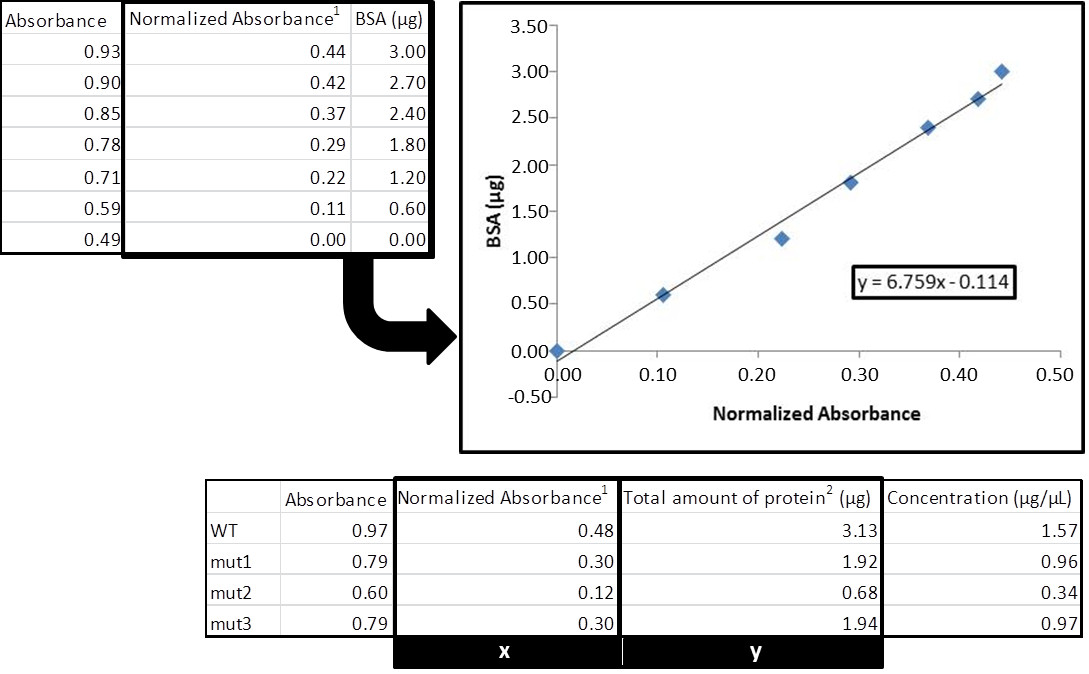
Figure 2. Total protein quantification with Bradford assay. 1Normalized absorbance is calculate by subtracting the absorbance value of the blank from the value obtained for each sample. 2Divide this value by the volume of protein extract used in the assay to calculate the final concentration (in this case, we used 2 μl).
- Figure 2 shows an example of protein quantification using the Bradford method.
- Prepare the BSA standards for the calibration curve as follows:
- HCT activity assay
HCT activity is measured by the conversion of p-coumaroyl-CoA and shikimate into p-coumaroyl shikimate (Figure 1).
- Prior to the preparation of enzymatic reactions, boil an aliquot of the xylem protein extract for 10 min, because the boiled protein extract will be used as negative control.
- The reaction mix is prepared in 1.5-ml tubes and contains 100 mM Tris-HCl pH 7, 1 mM DTT, 100 μM p-coumaroyl-CoA, 100 μM shikimic acid and 10 μg xylem protein extract (Recipe 2).
- Start the reaction by adding the corresponding volume of the protein extract. Use the same amount of protein of the boiled extract as negative control.
- Incubate at 30 °C for 30 min.
Note: For an in-depth analysis, different time-points can be used.
- Terminate the reaction by boiling the samples for 5 min.
Note: After boiling, place the reaction tubes on ice for at least 5 min, followed by a fast spin. This brings the droplets at the lid, caused by evaporation and condensation, back into the main sample, avoiding a change in the final product concentration.
- Transfer the total reaction volume (40 μl) to a LC sample vial for analysis of reaction products.
- Prior to the preparation of enzymatic reactions, boil an aliquot of the xylem protein extract for 10 min, because the boiled protein extract will be used as negative control.
- Product identification and quantification
10 μl of the aqueous phase is subjected to reversed phase LC-MS and LC-MS (Hoffmann et al., 2003). Here, the specific conditions can differ depending on the equipment used. We present our in house conditions optimized to identify and quantify p-coumaroyl shikimate on a Waters Acquity UPLC system connected to a Thermo LTQ XL mass spectrometer. Chromatographic separation is performed on an Acquity BEH C18 column (2.1 mm x 150 mm, 1.7 μm) using a gradient elution.
- The mobile phase is composed of water containing 1% acetonitrile and 0.1% formic acid (Buffer A) and acetonitrile containing 1% water and 0.1% formic acid (Buffer B). The column temperature is maintained at 40 °C and the autosampler temperature at 10 °C.
- A flow rate of 350 μl/min is applied during the gradient elution initializing at time 0 min 5% (B), time 30 min 50% (B), time 33 min 100% (B).
- The eluent is subsequently directed to the mass spectrometer, via electrospray ionization (ESI) in negative mode.
- MS source parameters are as follows: capillary temperature 300 °C, capillary voltage –24 V, source voltage 3.5 V, source current 100 μA, sheath gas flow 30, aux gas flow 20, sweep gas flow 5. The mass range is set between m/z 95-500.
- The reaction product, p-coumaroyl shikimate, is characterized based on m/z 319, retention times 8.13 min (trans isomer) 9.64 min (cis isomer), and fragmentation spectra (Figure 3). Compound concentrations were quantified based on curve fitting and peak area integration of the extracted ion chromatograms for m/z 319 using XCalibur's QuanBrowser.
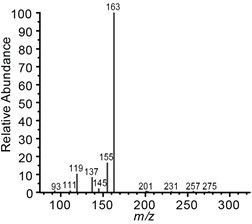
Figure 3. MS2 fragmentation spectra of p-coumaroyl shikimate (m/z 319)
Note: For accurate mass measurements of p-coumaroyl shikimate, a Synapt HDMS Q-Tof mass spectrometer can be used (Vanholme et al., 2013b).
- The mobile phase is composed of water containing 1% acetonitrile and 0.1% formic acid (Buffer A) and acetonitrile containing 1% water and 0.1% formic acid (Buffer B). The column temperature is maintained at 40 °C and the autosampler temperature at 10 °C.
Recipes
- Protein Extraction Buffer
Prepare the protein extraction buffer as follows:
Mix thoroughly.Stock Solution
Volume/Amount
Final Concentration
100 mM Tris-HCl pH 7.5
2 ml
20 mM
100 mM DTT
1 ml
10 mM
100% Glycerol
1.5 ml
15%
PVPP
100 mg
1%
7x Complete Mini Protease Inhibitor
1.33 ml
1x
WaterTo
10 ml
Notes:- Dissolve one tablet of Complete Mini Protease Inhibitor in 1.5 ml water to prepare the 7x stock solution.
- Since PVPP is insoluble, it may be necessary to cut the end of the pipet tip to add the protein extraction buffer to the ground xylem material.
- Dissolve one tablet of Complete Mini Protease Inhibitor in 1.5 ml water to prepare the 7x stock solution.
- Reaction Mix
Prepare the enzymatic reaction mix as follows:Stock Solution
Volume
Final Concentration
500 mM Tris-HCl pH 7.0
8 μl
100 mM
20 mM DTT
2 μl
1 mM
2 mM p-coumaroyl-CoA
2 μl
100 μM
2 mM shikimic acid
2 μl
100 μM
Xylem protein extract
x μl
10 μg
WaterTo
40 μl
Acknowledgments
The protocol was briefly described by Vanholme and coworkers in Vanholme et al. (2013a). We gratefully acknowledge funding through the European Commission’s Directorate-General for Research within the 7th Framework Program (FP7/2007-2013) under the grant agreement N° 211917 (ENERGYPOPLAR), N° 211868 (NOVELTREE) and N° 311804 (MULTIBIOPRO), the Hercules program of Ghent University for the Synapt Q-Tof (grant no. AUGE/014) and the Multidisciplinary Research Partnership ‘Biotechnology for a Sustainable Economy’ (01MRB510W) of Ghent University. RV is indebted to the Research Foundation-Flanders for a postdoctoral fellowship.
References
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Hoffmann, L., Maury, S., Martz, F., Geoffroy, P. and Legrand, M. (2003). Purification, cloning, and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem 278(1): 95-103.
- Hoffmann, L., Besseau, S., Geoffroy, P., Ritzenthaler, C., Meyer, D., Lapierre, C., Pollet, B. and Legrand, M. (2004). Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16(6): 1446-1465.
- Vanholme, B., Cesarino, I., Goeminne, G., Kim, H., Marroni, F., Van Acker, R., Vanholme, R., Morreel, K., Ivens, B., Pinosio, S., Morgante, M., Ralph, J., Bastien, C. and Boerjan, W. (2013a). Breeding with rare defective alleles (BRDA): a natural Populus nigra HCT mutant with modified lignin as a case study. New Phytol 198(3): 765-776.
- Vanholme, R., Cesarino, I., Rataj, K., Xiao, Y., Sundin, L., Goeminne, G., Kim, H., Cross, J., Morreel, K., Araujo, P., Welsh, L., Haustraete, J., McClellan, C., Vanholme, B., Ralph, J., Simpson, G.G., Halpin, C. and Boerjan, W. (2013b). Caffeoyl Shikimate Esterase (CSE) Is an Enzyme in the Lignin Biosynthetic Pathway. Science DOI: 10.1126/science.1241602.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cesarino, I., Vanholme, R., Goeminne, G., Vanholme, B. and Boerjan, W. (2013). Shikimate Hydroxycinnamoyl Transferase (HCT) Activity Assays in Populus nigra. Bio-protocol 3(22): e978. DOI: 10.21769/BioProtoc.978.
Category
Plant Science > Plant biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










