- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Visualization and Quantification of Actin Dynamics in Rice Protoplasts
Published: Vol 3, Iss 21, Nov 5, 2013 DOI: 10.21769/BioProtoc.964 Views: 9619
Reviewed by: Tie Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2322 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1690 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 756 Views
Abstract
Direct visualization of the organization and dynamics of the actin cytoskeleton in rice cells is essential to understand its roles in regulating rice growth and development. Visualization of actin dynamics in protoplasts transformed with actin probe is a relatively quick and simple strategy, compared to the strategy of generating stable transgenic rice plants that harbor actin probe, which is time-consuming. Here is a protocol described in details regarding transforming rice protoplasts as well as visualizing and quantifying actin dynamics in rice protoplasts, which is based on the method previously reported (Shi et al., 2013).
Keywords: Rice protoplastMaterials and Reagents
- Rice protoplasts
- Plasmid pUN1301-EGFP-ABD2-EGFP (Yang et al., 2011)
- D-Mannitol (Sigma-Aldrich, catalog number: M1902 )
- 2-(N-Morpholino) ethanesulfonic acid (MES) (Merck KGaA, catalog number: 475893 )
- Cellulase “Onozuka” RS (Yakult Pharmaceutical Industry, catalog number: 9012-54-8 )
- Macerozyme R-10 (Yakult Pharmaceutical Industry, catalog number: 8032-75-1 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A1933 )
- Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: C5670 )
- β-mercaptoethanol (Merck KGaA, catalog number: 444203 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S5886 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P5405 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M2393 )
- PEG4000 (Merck KGaA, catalog number: 817006 )
- Murashige and Skoog basal salt mixture powder (MS) (Sigma-Aldrich, catalog number: M5524 )
- Digestion buffer (see Recipes)
- W5 buffer (see Recipes)
- Mmg buffer (see Recipes)
- Polyethylene glycol solution (see Recipes)
Equipment
- 35 μm nylon mesh
- Spinning disk confocal microscope equipped with a Yokogawa CSU-X1 spinning disk head using a 512 x 512 Andor iXON electron-multiplying CCD camera (Andor)
- Shaker
- Centrifuge
- Concave slide (diameter 15 mm) (Beyotime, catalog number: FSL011 )
- Olympus BX61 inverted microscope equipped with a 100x 1.4 NA UPLSAPO (Universal Plan Super Apochromat Objective) objective
- 488 nm laser
- 525/50 nm band-pass filter
Software
- Andor IQ2 software (available at http://www.andor.com)
Procedure
- Sterilize rice seeds with 2.5% NaClO and 0.01% Tween-20 for 15 min and wash the seeds with sterilized water for five times, then sterilize the seeds with 2.5% NaClO for another 15 min and wash the seeds with sterilized water for five times. Finally put these seeds on sterilized filter paper and dry them.
- Germinate and grow sterilized rice seeds above on half-strength MS medium at 28 °C for 14 d in darkness.
- Cut the leaf sheaths of the etiolated seedlings into 0.5-1.0 mm length and immediately immerge into digestion buffer to digest the tissues in darkness at 28 °C for 4 h with gentle shaking (40-80 rpm). All steps below are operated at room temperature unless otherwise noted.
- Add one volume of pre-cooling W5 buffer into above mixture and collect the protoplasts on ice by sifting out the undigested tissues with 35 μm nylon mesh.
- Collect the protoplasts by a 5 min spinning of 300 x g at 4 °C and re-suspend the protoplasts in fresh cool W5 buffer.
- Harvest the pellet and dilute the protoplasts into 2 x 106 cells/ml with Mmg buffer.
- For transformation, 20 μg of plasmid pUN1301-EGFP-ABD2-EGFP was added into 100 μl protoplasts, followed by the addition of an equal volume of polyethylene glycol solution. The mixture was incubated at room temperature for 20 min.
- Add 10 volume of W5 buffer and harvest the protoplasts with centrifugation at 300 x g for 5 min.
- Re-suspend the pellet with fresh W5 buffer and incubate the protoplasts in darkness for 20 h.
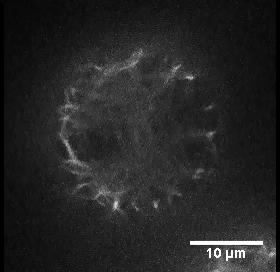
- Mount the protoplasts into a concave slide (diameter 15 mm) with a pipet and cover them with a coverslip. The dynamics of actin filaments were observed under an Olympus BX61 inverted microscope equipped with a 100x 1.4 NA UPLSAPO objective. Figure 1 shows the organization of actin filaments in a typical rice protoplast.

Figure 1. Organization of actin filaments in a typical rice protoplast. This protoplast is transformed with plasmid pUN1301-EGFP-ABD2-EGFP. Scale bar = 10 μm. - The images were collected using a spinning disk confocal microscope equipped with a Yokogawa CSU-X1 spinning disk head using a 512 x 512 Andor iXON electron-multiplying CCD camera. GFP was excited using a 488 nm laser, and fluorescence emission was collected using a 525/50 nm band-pass filter. Time-lapse Z-series images (with a step of 0.5 μm) were collected with a time interval of 100 msec using Andor IQ2, and the time interval for the whole Z-series collection was 5 sec. The Z-stack image is made into a movie to shown the actin filaments in a typical rice protoplast (see Video 1).
Video 1. A video of the Z-stack image displays the actin filaments in a typical rice protoplast harboring pUN1301-EGFP-ABD2-EGFP. Images were taken every 0.5 μm and were compressed into a 2.8 s-video. Scale bar = 10 μm. - The severing frequency of filament is quantified as the number of breaks, per unit filament length, per unit time (breaks/μm/s) and the depolymerization rate is calculated by the length of shortening filament within the given time interval (μm/s). Table 1 makes an example for one of our results.
Table 1. The severing frequency and depolymerization rate of filaments in rice protoplasts
Quantification of parameters associated with single actin dynamics in rice protoplasts harboring plasmid pUN1301-EGFP-ABD2-EGFP. More than 15 protoplasts were used to analyze. Values given are means± SE.Severing frequency (breaks/μm/s) 0.0165 ± 0.0080 (n = 15) Depolymerization rate (μm/s) 0.0023 ± 6.58e-4 (n = 20)
Recipes
- Digestion buffer
0.6 M mannitol
10 mM MES pH 5.7
1.5% cellulose RS
0.75% macerozyme R10
0.1% BSA
1 mM CaCl2
5 mM β-mercaptoethanol - W5 buffer
154 mM NaCl
125 mM CaCl2
5 mM KCl
2 mM MES - Mmg buffer
0.6 M mannitol
15 mM MgCl2
4 mM MES - Polyethylene glycol solution
0.6 M mannitol
100 mM CaCl2
40% v/v PEG 4,000
Acknowledgments
This protocol was adapted from our previously published work (Shi et al., 2013). We thank Meng Shi and Shaojie Cui for helpful suggestions on rice protoplast preparation and transformation. The research in the Huang lab was supported by grants from Ministry of Science of Technology (2013CB945100) and National Natural Science Foundation of China (31125004).
References
- Shi, M., Xie, Y., Zheng, Y., Wang, J., Su, Y., Yang, Q. and Huang, S. (2013). Oryza sativa actin-interacting protein 1 is required for rice growth by promoting actin turnover. Plant J 73(5): 747-760.
- Yang, W., Ren, S., Zhang, X., Gao, M., Ye, S., Qi, Y., Zheng, Y., Wang, J., Zeng, L., Li, Q., Huang, S. and He, Z. (2011). BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 23(2): 661-680.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Xie, Y. and Huang, S. (2013). Visualization and Quantification of Actin Dynamics in Rice Protoplasts. Bio-protocol 3(21): e964. DOI: 10.21769/BioProtoc.964.
Category
Plant Science > Plant cell biology > Cell imaging
Cell Biology > Cell imaging > Confocal microscopy
Plant Science > Plant cell biology > Cell structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link