- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Immunoplaque Assay (Influenza Virus)
Published: Vol 3, Iss 21, Nov 5, 2013 DOI: 10.21769/BioProtoc.959 Views: 25760
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Image-based Quantification of Direct Cell-to-cell Transmission of Bovine Viral Diarrhea Virus
Fernando Merwaiss and Diego E. Alvarez
Aug 5, 2019 5323 Views

An Improved Focus-Forming Assay for Determination of the Dengue Virus Titer
Maharah Binte Abdul Mahid [...] Kitti Wing Ki Chan
Oct 20, 2024 2415 Views
Abstract
Despite developed long time ago, plaque assay is still the gold standard for viral titer quantification in modern virology. The standard crystal violet-based plaque assay relies on virus’ ability to induce cytopathic effect (CPE) which limits the assay to lytic viruses. Alternative viral quantification assays such as 50% tissue culture infectious assay (TCID50) and genetic material quantification by Q-PCR provide a different way of viral quantification with their own shortcoming. In here, we modified the fluorescent focus assay and developed an antibody-based immunoplaque assay which provides a reliable and reproducible viral quantification independent of CPE. Our assay not only allows accurate determination of viral titer, but also provides information on viral kinetics, genetic stability and purity of the virus population.
Keywords: InfluenzaMaterials and Reagents
- MDCK cells
Please note that there are multiple variants of MDCK on American Type Culture Collection (ATCC). This protocol has been tested for MDCK (CCL-34), MDCK.1 (CRL-2935) and MDCK.2 (CRL-2936)
- Recombinant A/California/04/2009 virus produced from 12 plasmids reverse genetic system (Generous gift from Professor Toru Takimoto from University of Rochester, New York, USA) (Bussey et al., 2010)
- Trypsin/EDTA (Mediatech, Cellgro®, catalog number: 25-053-CI )
- SeaPlaque Agarose (Lonza, catalog number: 50100 )
- TPCK-Trypsin (Thermo Fisher Scientific, catalog number: 20233 )
- PBS without Ca2+ and Mg2+ (Mediatech, Cellgro®, catalog number: 21-030-CV )
- DPBS with Ca2+ and Mg2+ (Mediatech, Cellgro®, catalog number: 21-040-CV )
- 10% formalin (Sigma-Aldrich, catalog number: HT501128-4L )
- TritonX-100 (Sigma-Aldrich, catalog number: T9284 )
- 5% goat serum in DPBS (Gibco, catalog number: 16210-072 )
- Mouse anti-NP antibody from hybridoma H16-L10-4R5 (ATCC, catalog number: HB-65 )
- Alkaline phosphatase conjugated Donkey anti-mouse antibody (Jackson ImmunoResearch Laboratories, catalog number: 715-056-150 )
- Alexa Fluor 488 conjugated Goat anti-mouse antibody (Invitrogen, catalog number: A11001 )
- BCIP/NBT substrate (Vector Laboratories, catalog number: SK-5400 )
- DMEM (Mediatech, Cellgro®, catalog number: 10-017-CV )
- DMEM powder (Mediatech, Cellgro®, catalog number: 50-003-PB )
- Heat-inactivated fetal bovine serum (Mediatech, Cellgro®, catalog number: 26140079 )
- HEPES (Mediatech, Cellgro®, catalog number: 25-060-CI )
- Penicillin/Streptomycin (Mediatech, Cellgro®, 30-002-CI )
- 7.5% BSA Fraction V (Gibco, catalog number: 15260 )
- RPMI 1640 powder (Mediatech, Cellgro®, catalog number: 50-020-PB )
- Tris base (US Biological, catalog number: T8600 )
- Sodium chloride (NaCl)
- Magnesium chloride (MgCl2)
- Sodium hydroxide (NaOH)
- Complete DMEM (cDMEM) (see Recipes)
- 2x DMEM (see Recipes)
- RPMI (see Recipes)
- 0.5% Triton X-100 in DPBS (see Recipes)
- Alkaline phosphatase (AP) reaction buffer (see Recipes)
Equipment
- Standard tissue culture equipment
- 12-well tissue culture plate
- 37 °C, 5% CO2 cell culture incubator
- Inverted fluorescence microscope (Optional)
- Rocker
- Laboratory spatula
- Heated stir plate
- Vortex
- Axiovert 200 microscope
Software
- IPlab3.6.5 software
Procedure
Day 1
- Each well on a 12-well plate is seeded with 5 x 105 MDCK cells suspended in 1 ml of cDMEM.
- The plate is incubated in the 5% CO2 incubator at 37 °C overnight.
Day 2 (or until cells reach 95% - 100% confluence)
- Viruses are serially diluted (10-fold) in RPMI, vortex each dilutions and keep on ice before use.
- MDCK cells are rinsed with 1 ml of DPBS twice.
- MDCK cells are inoculated with 500 μl of viruses prepared from step 3.
- The plate is incubated in 5% CO2 incubator at 37 °C for 45 min.
- Prepare SeaPlaque DMEM (pDMEM)
- 2x DMEM is warmed at 37 °C water bath and 2% SeqPlaque agarose is melted on a hot plate at about 45–50 °C with stirring (*The cap should be slightly released to prevent pressure buildup).
- To obtain pDMEM, pre-warm 2x DMEM and 2% agarose are mixed in 1:1 ratio in a sterile container and sit at room temperature for 5–10 min to cool down.
- TPCK-trypsin (final concentration: 1 μg/ml) is added to the pDMEM.
- 2x DMEM is warmed at 37 °C water bath and 2% SeqPlaque agarose is melted on a hot plate at about 45–50 °C with stirring (*The cap should be slightly released to prevent pressure buildup).
- Viral inoculums are aspirated and MDCK cells are rinsed with 1 ml of DPBS once.
- 1.5 ml pDMEM + TPCK-trypsin from step 7-c is added to each well.
- The plate is sat at room temperature (RT) for 10–15 min until pDMEM solidify.
- The plate is incubated at 5% CO2 at 37 °C in the invert orientation (lid at the bottom) for 3 days.
Day 5 (or until visualization of plaque)
- MDCK cells are fixed by adding 1.5 ml of 10% formalin directly on top of the agarose and incubated at RT for 2 h.
- Extra formalin is aspirated and agarose from the well is removed and trashed carefully without disturbing the monolayer of cell using a laboratory spatula or equivalent tools.
Note: Experiment can be stopped after this step by adding PBS to cover the cells and store at 4 °C.
- MDCK cells are permeabilized by 300 μl of 0.5% Triton-X100 in DPBS for 5 min on bench without agitation.
- Cells are rinsed with 1 ml of PBS twice.
- Cells are blocked by 300 μl of 5% goat serum in DPBS for 30 min at RT with agitation on rocker.
- Cells are incubated with H16-L10-4R5 antibody (1:40) in 300 μl of 5% goat serum for 1 h at RT with agitation on a rocker.
- Cells are rinsed with 1 ml of PBS twice.
- Cells are incubated with alkaline phosphatase conjugated donkey anti-mouse (1:1,000) in 300 μl of 5% goat serum for 45 min at RT with agitation on a rocker.
- Cells are rinsed with 1 ml of PBS twice and AP reaction buffer once.
- Prepare the BCIP/NBT solution.
- Added 2 drops of solution A, B and C into every 5 ml of AP reaction buffer.
- Mix by inverting the tube for 4–6 times.
- Added 2 drops of solution A, B and C into every 5 ml of AP reaction buffer.
- 300 μl of BCIP/NBT solution is added to each well. The plate is put on a rocker in dark for 30 min or until the development of color.
- The reaction is stopped by rinsing the plate with ddH2O.
- The plate is air-dried at RT.
- Count the number of plaques, check the plaque morphology and measure the size of plaques.
Alternative visualization method by fluorescence, fluorescent focus assay (FFA)
(Continuous from step 18)
- 19.1
- Cells are incubated with goat anti-mouse Alexa Fluor 488 antibody (1:1,000) in 300 μl of 5% goat serum for 45 min at RT in dark.
21.1 Cells are covered with 500 μl of DPBS and store at 4 °C.
- 22.1
- Check the plaque morphology and measure the size of plaques using an inverted fluorescence microscope (Figure 1).
Note: The protocol is optimized for influenza viruses. Modifications may be required for use in other viruses. For instance, factors required for multiple rounds of replication of the virus should be added in the pDMEM.
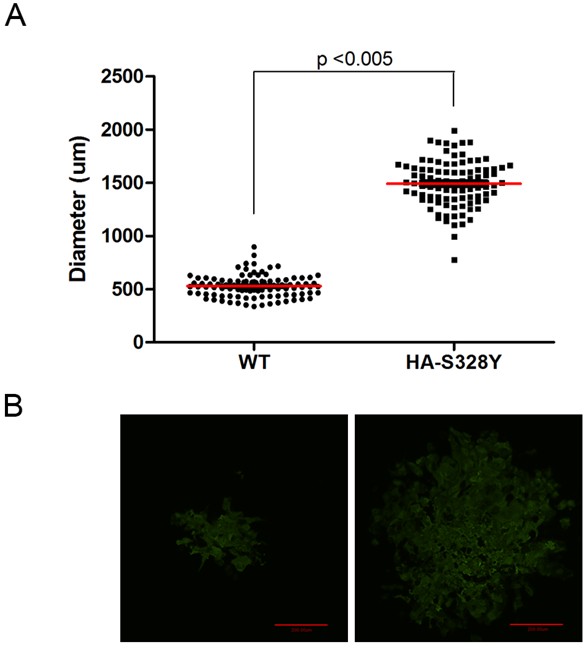
Figure 1. Diameter of Individual Plaque of A/California/04/2009/H1N1 Recombinatant Viruses (rCA0409). A. Quantification of the diameter (μm) of 100 individual plaques of two different CA0409 viruses, wild type (WT) and HA-S328Y mutants (HA328Y). Images are taken by Axiovert 200 microscope using a 5x objective and diameters of each plaque are measured by IPlab3.6.5 software. Statistics are done by one-tail Student’s t-test. B. Representative image of plaques of CA0409 viruses stained with Alexa Fluor 488, scale bar = 200 μm. Images are taken by Sensicam using IPlab 3.6.5 (Data not published) (Tse et al., 2013).
Recipes
- cDMEM (filtered in 0.2 μm filtration unit)
DMEM
432.5 ml
Heat-inactivated fetal bovine serum
50 ml
1 M HEPES
12.5 ml
Penicillin/Streptomycin
5 ml
- 2x DMEM (filtered in 0.2 μm filtration unit)
DMEM powder
13.48 g
7.5% BSA Fraction V
26.7 ml
1 M HEPES
12.5 ml
Penicillin/Streptomycin
5 ml
ddH2O to 500 ml
- RPMI (filtered in 0.2 μm filtration unit)
RPMI 1640 powder
5.2 g
7.5% BSA fraction V
13.3 ml
1 M HEPES
12.5 ml
Penicillin/Streptomycin
5 ml
ddH2O to 500 ml
- 0.5% Triton X-100 in DPBS
Triton X- 100
1 ml
DPBS
199 ml
- AP reaction buffer
Tris
100 mM
NaCl
100 mM
MgCl2
5 mM
Adjust pH to 9.5 using NaOH
Acknowledgments
We thank Jean K. Millet, Tamar Friling, Alice M. Hamilton and all of the members of the Whittaker lab for helpful discussions. We also thank the Collins lab for helpful suggestions made throughout the study.
References
- Bussey, K. A., Bousse, T. L., Desmet, E. A., Kim, B. and Takimoto, T. (2010). PB2 residue 271 plays a key role in enhanced polymerase activity of Influenza A viruses in mammalian host cells. J Virol 84(9): 4395-4406.
- Tse, L. V., Marcano, V. C., Huang, W., Pocwierz, M. S. and Whittaker, G. R. (2013). Plasmin-mediated activation of pandemic H1N1 influenza virus hemagglutinin is independent of the viral neuraminidase. J Virol 87(9): 5161-5169.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tse, L. V., Zhang, Y. and Whittaker, G. R. (2013). Immunoplaque Assay (Influenza Virus). Bio-protocol 3(21): e959. DOI: 10.21769/BioProtoc.959.
Category
Microbiology > Microbial cell biology > Cell isolation and culture
Cell Biology > Cell imaging > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link