- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Surface Polysaccharide Extraction and Quantification
Published: Vol 3, Iss 20, Oct 20, 2013 DOI: 10.21769/BioProtoc.934 Views: 19616
Reviewed by: Fanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Simple Methods for the Preparation of Colloidal Chitin, Cell Free Supernatant and Estimation of Laminarinase
Ananthamurthy Koteshwara
Oct 5, 2021 5163 Views

Extraction and Electrophoretic Analysis of Bacterial Lipopolysaccharides and Outer Membrane Proteins
Yue-Jia Lee and Thomas J. Inzana
Dec 20, 2021 5757 Views

Surface Plasmon Resonance for the Interaction of Capsular Polysaccharide (CPS) With KpACE
Zhe Wang [...] Chao Cai
Jun 20, 2025 3553 Views
Abstract
Gram-negative bacterial cells possess two membranes - the inner cytoplasmic membrane and the outer membrane. The two membranes are distinct in their composition; the inner membrane is composed of a phospholipid bilayer, whereas the outer membrane (OM) is composed of an asymmetrical bilayer, with the outer leaflet containing lipopolysaccharide (LPS) (Raetz and Whitfield, 2002). Surface polysaccharides, such as LPS O-antigen, or capsular polysaccharide, are often tightly associated with the OM (Whitfield, 2006). This tight association can be used to generate a rough quantification of surface polysaccharides of Gram-negative bacterial cells, as the OM can easily be dissociated from cells without associated cell lysis (Brimacombe et al., 2013). The following method describes how to quickly extract and quantify OM-associated polysaccharides.
Keywords: PolysaccharideMaterials and Reagents
- Culture of bacterial cells (This procedure works only for Gram-negative bacteria, for example Escherichia coli, Pseudomonas aeurginosa, or Rhodobacter capsulatus. The outer membrane, specifically LPS, is essential for this procedure to work)
- 50 mM sodium chloride (NaCl) dissolved in deionized H2O
- 50 mM ethylenediaminetetraacetic acid (EDTA) (EMD Millipore, catalog number: 324503 )
- Phenol (Fisher Scientific, catalog number: A92-100 )
- 93% sulfuric acid (Avantor Performance Materials, catalog number: 2900-10 )
- Carbohydrate stock solution (see Recipes)
Equipment
- Microcentrifuge
- Microfuge tubes (ESBE, catalog number: ESB-ES00507C )
- Spectrophotometer
- Cuvettes
- Glass test tubes
- Glass pipettes
Procedure
- Extraction of surface polysaccharides from Gram-negative bacteria
- Grow bacteria to desired growth phase (generally stationary phase) in desired growth medium.
Note: Growth medium may affect surface polysaccharide levels, so the same media should be used for all experiments if possible. - Measure OD650 of culture; dilute to < 1 OD if necessary to get an accurate measurement.
- Normalize cultures to OD650 of 2.0 (or to maximum OD that bacterial culture will grow to if it is less than 2.0).
- Harvest 1 ml of each normalized culture by centrifugation at 14,500 x g for 5 minutes in a microcentrifuge.
- Carefully remove supernatant with a pipette, discard tip.
- Wash cells by re-suspending in 1 ml of 50 mM NaCl, pellet by centrifugation at 14, 500 x g for 5 minutes, remove supernatant.
- Repeat step A6 four additional times (5 total washes).
- Re-suspend cells in 1 ml of 50 mM EDTA, and incubate at 37 °C for 60 minutes (EDTA causes LPS to dissociate, thus releasing the OM from cells).
- Pellet cells by centrifugation at 14,500 x g for 5 minutes, carefully remove supernatant and transfer to fresh microfuge tube (supernatant contains all surface polysaccharides, including LPS, capsule etc.).
- Grow bacteria to desired growth phase (generally stationary phase) in desired growth medium.
- Quantification of surface polysaccharides
- Prepare carbohydrate standards by diluting carbohydrate stock solution into 1 ml aliquots of: 0, 30, 60, 90, and 120 μg/ml of carbohydrate (e.g. 970 μl of dH2O + 30 μl of 1 mg/ml stock solution to generate a 30 μg/ml standard).
- Prepare clean, acid washed glass test tubes (for a suggested protocol, see Reference 4). Pipette 200 μl of standards, a 200 μl control of 50 mM EDTA, and all test samples into separate tubes.
- Move to fume hood.
- Add 200 μl of 5% phenol to all tubes, mix well by shaking.
- Add 1 ml of 93% sulfuric acid; mix well by swirling (use caution).
- Allow colour to develop for 10 minutes at room temperature (reaction should turn yellow; intensity depends on carbohydrate concentration). Additional mixing by gentle swirling every 2-3 minutes may help reaction proceed faster.
- Measure OD490 of all reactions in a spectrophotometer; concentration of carbohydrates can then be calculated from the standard curve.
Note: If necessary, dilute reactions in dH2O to get accurate spectrophotometer readings.
- Prepare carbohydrate standards by diluting carbohydrate stock solution into 1 ml aliquots of: 0, 30, 60, 90, and 120 μg/ml of carbohydrate (e.g. 970 μl of dH2O + 30 μl of 1 mg/ml stock solution to generate a 30 μg/ml standard).
Representative data
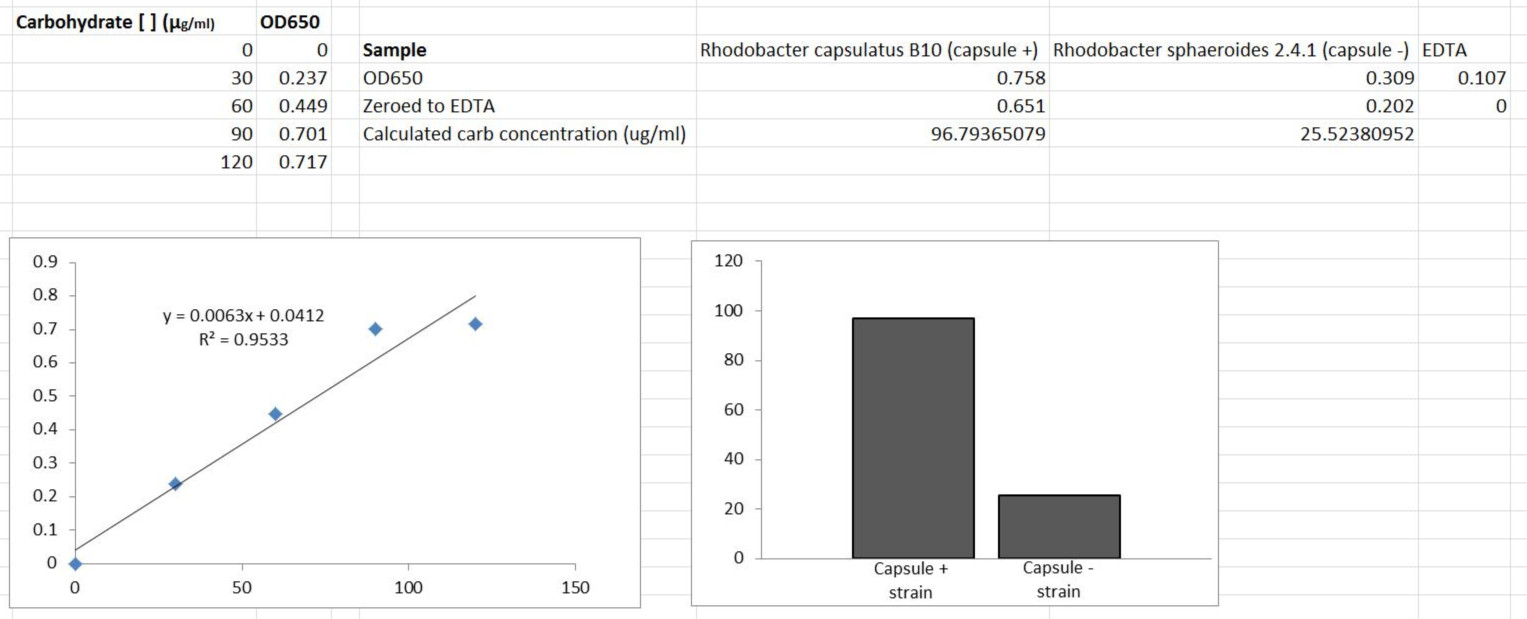
Recipes
- Carbohydrate stock solution
50:50 mixture of 0.5 mg/ml each of sucrose and fructose
Final concentration of 1 mg/ml carbohydrate (molecular biology grade recommended)
Acknowledgments
The development of this protocol was funded by a grant to J.T.B. from the Canadian Institutes of Health Research.
References
- Brimacombe, C. A., Stevens, A., Jun, D., Mercer, R., Lang, A. S. and Beatty, J. T. (2013). Quorum-sensing regulation of a capsular polysaccharide receptor for the Rhodobacter capsulatus gene transfer agent (RcGTA). Mol Microbiol 87(4): 802-817.
- http://openwetware.org/images/9/9e/GLASSWARE_CLEANING_PROCEDURES.pdf
- Raetz, C. R. and Whitfield, C. (2002). Lipopolysaccharide endotoxins. Annu Rev Biochem 71: 635-700.
- Whitfield, C. (2006). Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem 75: 39-68.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Brimacombe, C. A. and Beatty, J. T. (2013). Surface Polysaccharide Extraction and Quantification. Bio-protocol 3(20): e934. DOI: 10.21769/BioProtoc.934.
Category
Microbiology > Microbial biochemistry > Carbohydrate
Biochemistry > Carbohydrate > Polysaccharide
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










