- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro T Cell–DC and T Cell–T Cell Clustering Assays
Published: Vol 3, Iss 20, Oct 20, 2013 DOI: 10.21769/BioProtoc.933 Views: 15811
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Functional Phenotyping of Lung Mouse CD4+ T Cells Using Multiparametric Flow Cytometry Analysis
Céline M. Maquet [...] Bénédicte D. Machiels
Sep 20, 2023 4206 Views

T-Cell-Based Platform for Functional Screening of T-Cell Receptors Identified in Single-Cell RNA Sequencing Data Sets of Tumor-Infiltrating T-Cells
Aaron Rodriguez Ehrenfried [...] Rienk Offringa
Apr 20, 2024 6409 Views

Novel Experimental Approach to Investigate Immune Control of Vascular Function: Co-culture of Murine Aortas With T Lymphocytes or Macrophages
Taylor C. Kress [...] Eric J. Belin de Chantemèle
Sep 5, 2025 3551 Views
Abstract
To get activated, T cells need to find their cognate antigen at the surface of an antigen-presenting cell (APC). Recognition of cognate antigen in the context of the MHC (Major histocompatibility complex) by the TCR (T-Cell Receptor) results in long lasting interactions between T cells and APCs. Subsequently, T cells form homotypic interactions with each other, which is seen as a hallmark of T cell activation. This protocol describes a method to analyze T-APC and T-T conjugation.
Materials and Reagents
- RPMI-1640 medium (Life Technologies, Gibco®, catalog number: 11875-093 )
- FBS (Hyclone, catalog number: SH30396-03 )
- Penicillin-Streptomycin-Glutamine Solution (Life Technologies, Gibco®, catalog number: 10378-016 )
- 2-mercaptoethanol (Life Technologies, Invitrogen™, catalog number: 21985-023 )
- Phorbol 12-myristate-13-acetate (PMA) (Sigma-Aldrich, catalog number: 79346 )
- Ionomycin (Sigma-Aldrich, catalog number: 19657 )
- 5-(and-6)-Carboxyfluorescein Diacetate, Succinimidyl Ester (5(6)-CFDA, SE) (CFSE) (Life Technologies, Molecular Probes®, catalog number: C1157 )
- 7-hydroxy-9H-(1,3-dichloro-9,9-dimethylacridin-2-one (DDAO) (Life Technologies, Invitrogen™, catalog number: C34553 )
- 5-(and-6)-(((4-Chloromethyl)Benzoyl)Amino) Tetramethylrhodamine (CMTMR) (Life Technologies, Invitrogen™, catalog number: C2927 )
- 16% Paraformaldehyde Solution (PFA) (16% Formaldehyde, EM Grade) (Electron Microscopy Sciences, catalog number: 15710 )
- Vectashield Hardset with Dapi (VWR International, catalog number: 101098-050 )
- Poly-L-Lysine Solution 0.1% (w/v) in H2O (Sigma-Aldrich, catalog number: P8920 )
- SL8 (Ovalbumin (257-264)) (AnaSpec, catalog number: 60193 )
- PBS (Life Technologies, catalog number: 14190-44 )
- Media (see Recipes)
- 2% PFA (see Recipes)
- Poly-L-Lysine-coated chamber (see Recipes)
Equipment
- Centrifuge
- Flow Cytometer
- Epifluorescent microscope
- 37 °C, 5% CO2 Cell culture incubator
- 24 well plate (Corning, Costar®, catalog number: 3524 )
- 40 μm Cell strainer (BD Biosciences, Falcon®, catalog number: 352340 )
- FACS tube, polystyrene (BD Biosciences, Falcon®, catalog number: 352054 )
- 8-well chamber slide (Thermo Fisher Scientific, Lab-TekTM, catalog number: 177445 )
Procedure
- T-APC conjugation assay
Notes:
- This protocol uses purified OT-I CD8+ T-cells from naive mice, which are transgenic for a TCR recognizing the chicken egg OVA–derived SIINFEKL peptide (SL8) in the context of the MHC class I molecule H2-kb. The same protocol can be used with T cells specific for other antigens. BMDCs (Bone-Marrow derived Dendritic Cells) are used as APCs, but DCs from other origin, or many cell lines, can be used as APCs.
- Once cells are labeled, they should stay protected from light as much as possible.
- Cell labelling: resuspend BMDCs and purified naive OT-I cells in PBS at a concentration of 10 x 106 cells/ml (usually 20 x 106 cells in 2 ml). Add DDAO at a final concentration of 4 μM on BMDCs, and CFSE at a final concentration of 1 μM on OT-I cells. Incubate for 30 min at 37 °C 5% CO2.
Note: CFSE and DDAO can be substituted for other dyes. The advantage using those 2 dyes is that little to no compensation on the flow cytometer is needed.
- Wash cells three times with 5 volumes of media (10 ml if you start with 20 x 106 cells) by centrifuging at 300 x g for 5 min at room temperature.
- Pulse APCs: resuspend BMDCs in media at a concentration of 10 x 106 cells/ml. Add SL8 at a final concentration ranging from 0-1,000 ng/ml (usually 5 different concentrations are used) and incubate for 60 min at 37 °C 5% CO2.
- While APCs are antigen pulsing, resuspend OT-I cells at 4 x 106 cells/ml in media (usually 20 x 106 cells in 5 ml). Add 50 μl of OT-I cells per FACS tube. Leave cells at 37 °C 5% CO2 until APCs are ready.
- Wash BMDCs three times with 5 volumes of media by centrifuging at 300 x g for 5 min at room temperature.
- Resuspend BMDCs at 4 x 106 cells/ml in media.
- Add 50 μl of BMDCs to each FACS tube containing OT-I cells. Immediately spin at 228 x g (1,000 rpm) for 1 min at 20 °C. Immediately place at 37 °C.
Note: In this protocol, the ratio T cell-APC is 1, but you can make it vary. Time of incubation is usually between 0-60 min (0-10-20-30-60 min).
- Fix the cells: Once the desired amount of time is completed, add 100 μl per sample of warm (20 °C) 2% PFA. Mix very gently, either by pipetting up and down or quickly vortexing on low. Incubate at room temperature (around 20 °C) for 10 min.
- Run on flow cytometer. Assess the percentage of coupling based on double CFSE/DDAO positives (Figure 1).
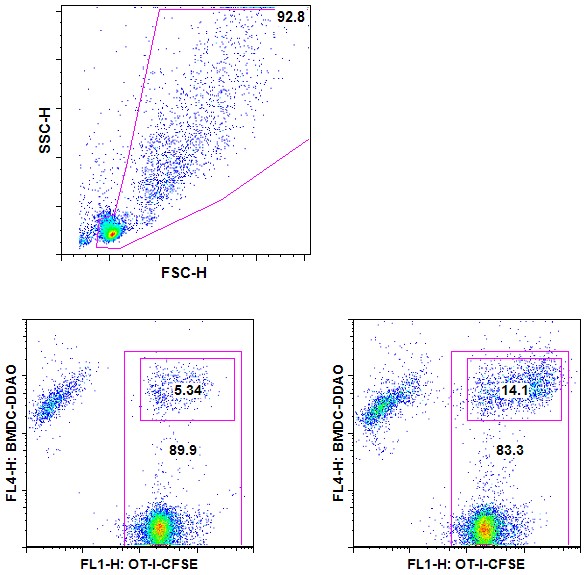
Figure 1. T-APC coupling assay-Example. CFSE-labeled OT-I cells are incubated for 30 min at 37 °C with DDAO-labeled BMDCs that were pulsed with 0 (left plot) or 100 ng/ml (right plot) SL8 peptide. Cells debris were first excluded using a SSC/FSC gate (upper panel). Coupled cells are DDAO+ and CFSE+.
- T-T clustering assay
Notes:
- In this protocol, T cells are activated with PMA and Ionomycin, which by-pass the need for an APC and every T cell could be activated this way. However, the same protocol can be used with T cells activated with an APC, anti-CD3 and anti-CD28, etc...
- This protocol compares, as an example, the capacity of OT-I cells deficient for the integrin ICAM-1 to form homotypic interactions compared to control OT-I cells. But the same protocol can be used to investigate the kinetic of T cell clustering, blocking antibodies, etc…
- Cell labelling: resuspend purified OT-I cells in PBS at a concentration of 10 x 106 cells/ml (usually 20 x 106 cells in 2 ml). Add CFSE at a final concentration of 1 μM on Control cells, and CMTMR at a final concentration of 4 μM on ICAM-1-/- cells. Incubate for 30 min at 37 °C 5% CO2.
- Wash cells three times with 5 volumes of media by centrifuging at 300 x g for 5 min at room temperature.
- T cell activation: Resuspend OT-I cells in media at a concentration of 2 x 106 cells/ml. WT and ICAM-1-/- cells are mixed at a ratio 1 and then activate cells by adding 5 ng/ml PMA and 50 ng/ml ionomycin directly in the media. Plate 2 ml cell suspension per well of a 24 well plate.
- 16 to 24 h after activation, percentage of cells in clustered is analyzed by flow cytometry (step B5) or microscopy (step B6).
- Analysis by flow cytometry: Cells are fixed by adding the same volume of 2% PFA for 10 min at 20 °C. Fixed cells are run through a 40 μm strainer into a FACS tube. Cells that went through represent the “non-clustered” fraction. Cells retained on the strainer are washed with 1 ml of PBS 2 mM EDTA (to disrupt cell clusters) and represent the “clustered” fraction. Both fractions are run on flow cytometer (Figure 2).
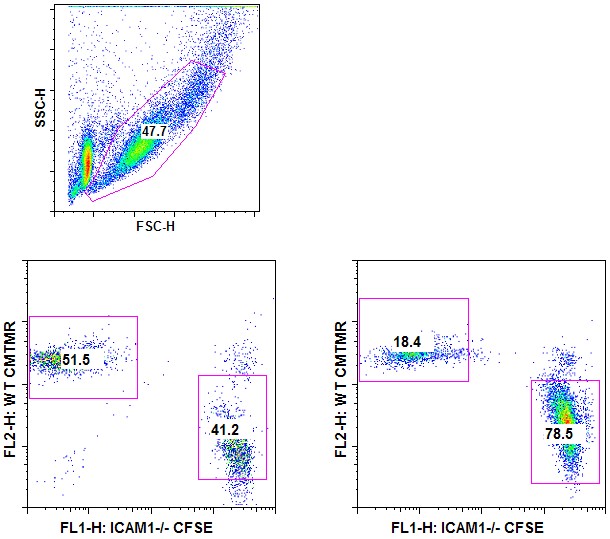
Figure 2. T-T clustering assay–Analysis by flow cytometry. CFSE-labeled WT OT-I cells and CMTMR-labeled ICAM-1-/- OT-I cells were ad-mixed and stimulated with PMA and Ionomycin. 24 h after activation, clustered cells were separated from non-clustered cells through a 40 μm strainer and both fractions were analyzed by flow cytometry. Cell debris was first excluded using a SSC/FSC gate (upper panel). Clustered cells – left panel; non-clustered cells – right panel.
Note that ICAM-1 deficient cells are mainly found in the non-clustered cell fraction compared to WT cells.
Note: Cut the tip of your pipet tip to ensure that cell clusters are not disrupted by pipetting when applying the cell suspension on the strainer.
In vitro activated T cells usually form clusters that are bigger than 40 μm diameter and therefore should be retained on the strainer. However, cells that are going through the strainer can potentially be part of smaller clusters. It is therefore useful to compare the flow cytometry data with microscopy quantification.
- Analysis by microscopy: Cells (usually 300 μl of cell suspension) are transferred onto a poly-L-Lysine-coated chamber and incubated for 10 min at 37 °C 5% CO2. Media is then carefully removed and replaced by 200 μl of 2% PFA. After 10 min, PFA is carefully removed, and replaced by 200 μl of PBS. Remove the PBS and let dry for 2 min. Remove the chamber gasket and mount with a drop of Vectashield on a coverslip. Analyze with epifluorescent microscope.
Note: Cut the tip of your pipet tip to ensure that cell clusters are not disrupted by pipetting when applying the cell suspension on the chamber.
Recipes
- Media (555 ml)
500 ml RPMI Medium 1640
50 ml heat inactivated (30 min at 56 °C) FCS
500 μl of 50 mM β-mercaptoethanol
5 ml of Penicillin-Streptomycin-Glutamine Solution
Keep at 4 °C
- 2% PFA
Dilute 1 ml of 16% PFA solution in 7 ml PBS
Keep in dark at 4 °C
- Poly-L-Lysine coated chambers
Dilute poly-L-Lysine stock solution 1/100 in dH2O
Put 500 μl on each chamber
Incubate 5-10 min at RT
Wash thoroughly with dH2O and let the chambers dry at least 2 h
Chambers can be stocked for several weeks at RT
Acknowledgments
This protocol was adapted from the previously published paper Gerard et al. (2013). This protocol was adapted in the laboratory of Matthew F. Krummel, and was supported by grants from the Juvenile Diabetes Foundation (MFK), and NIH R01AI52116 (MFK).
References
- Gérard, A., Khan, O., Beemiller, P., Oswald, E., Hu, J., Matloubian, M. and Krummel, M. F. (2013). Secondary T cell-T cell synaptic interactions drive the differentiation of protective CD8+ T cells. Nat Immunol 14(4): 356-363.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Gérard, A. (2013). In vitro T Cell–DC and T Cell–T Cell Clustering Assays. Bio-protocol 3(20): e933. DOI: 10.21769/BioProtoc.933.
Category
Immunology > Immune cell function > Lymphocyte
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









