- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Spindle Angle Measurements
Published: Vol 3, Iss 19, Oct 5, 2013 DOI: 10.21769/BioProtoc.925 Views: 12448
Reviewed by: Lin FangFanglian HeHui Zhu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An NMR Approach for Investigating Membrane Protein–Lipid Interactions Using Native Reverse Micelles
Sara H. Walters and Brian Fuglestad
Jul 20, 2024 2281 Views

A Protocol to Purify Human Mediator Complex From Freestyle 293-F Cells
Hui-Chi Tang [...] Ti-Chun Chao
Feb 20, 2025 2155 Views

Intraepidermal Nerve Fiber Quantification of the Mouse Hind Paw Footpads: A Detailed and Simplified Protocol
Anastasia Yerushkin [...] Amir Dori
Dec 5, 2025 1247 Views
Abstract
Spindle angles measures derive from the measures of spindle poles positions that were taken from fixed and immunostained adherent cells. To determine spindle angles (α), z-stack images of metaphasic cells immunostained with anti γ-tubulin (spindle poles) and anti β-tubulin antibodies (mitotic spindle) were acquired. A very simple ImageJ software macro was developed to measure the spindle angle using spindle pole coordinates (see Figure 1).
Keywords: Spindle
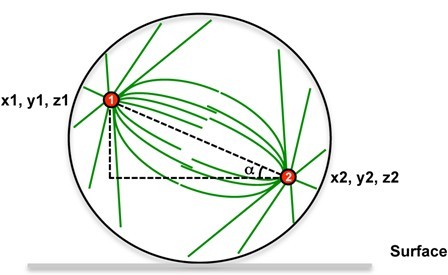
Figure 1. Spindle angle measurement principle. Spindle poles coordinates are measured, then the spindle angle alpha is calculated.
Materials and Reagents
- Fixed cells
- Antibodies
For example, for mitotic spindle poles, an antibody against γ-tubulin antibody (clone AK-15) (e.g. Sigma-Aldrich, catalog number: T3320 )
For mitotic spindle using an antibody directed against α-or β-tubulin (clone TUB 2.1) (e.g. Sigma-Aldrich, catalog number: T4026 )
Anti-rabbit coupled to Alexa Fluor® 555 (Life Technologies, Invitrogen™, catalog number: A21429 )
Anti-mouse coupled to Alexa Fluor® 488 (Life Technologies, Invitrogen™, catalog number: A11029 )
Note: Primary (AK-15 and TUB 2.1) and secondary (Anti-rabbit coupled to Alexa Fluor® 555 and anti-mouse coupled to Alexa Fluor® 488) antibodies were used after a 1,000 time dilution in 1x PBS/1% BSA.
- 10x PBS (Life Technologies, Invitrogen™, catalog number: AM9625 )
- 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma-Aldrich, catalog number: D9542 )
- BSA (Sigma-Aldrich, catalog number: A4503 )
- 1x PBS (see Recipes)
- 1x PBS/1% BSA (see Recipes)
Equipment
- We use a Zeiss Axioimager Z1 with 63x Plan-Apochromat 1.4 oil lens and using an Axiocam Mrm camera with a Grid Projection Illumination (apotome). The system is driven by Axiovision software. Images can also be obtained from any confocal microscopes/widefield microscope + deconvolution.
- 12 mm round coverslips (#1.5)
- Glass slides
- Vectashield® Mounting Media (Vector Laboratories, catalog number: H-1000 ) or ProLong® Gold (Life Technologies, Invitrogen™, catalog number: P36934 )
Software
- Axiovision software
- Open source software ImageJ 1.47q (http://rsbweb.nih.gov/ij/index.html)
- Macro for spindle angle measurements
Procedure
- Stain cells for spindle poles and mitotic spindle using the above-mentioned antibodies and according to the following protocol
- Cells grown on 12 mm round coverslips (#1.5) are fixed for 5 minutes at -20 °C in MeOH.
- Rehydrated in 1x PBS for 5 minutes and saturated in 1x PBS/1% BSA for at least 30 minutes at room temperature.
- Incubated for 2 hours at room temperature with primary antibodies in 1x PBS/1% BSA.
- After 3 washes of few seconds in 1x PBS/1% BSA, cells are incubated for 1 hour at room temperature with secondary antibodies coupled to fluorophores and DAPI (to stain DNA, DAPI was used at 0.5 μg/ml) in 1x PBS/1% BSA.
- After 3 washes in 1x PBS/1% BSA, coverslips are mounted onto glass slides using either Vectashield® or ProLong® Gold.
- Cells grown on 12 mm round coverslips (#1.5) are fixed for 5 minutes at -20 °C in MeOH.
- Imaging analysis
- Acquire Z stacks with a 63x/100x PLAN APO lens. Use Nyquist/Shannon criterion for Z step calculation (0.24 μm for this lens). Image quality must be good enough so that poles are clearly identified.
- Make sure the acquisition software calibrates the image (i.e. voxel size is included in the image Metadata). If not, the macro will request to calibrate the Image. XY pixel size can be derived from (physical camera pixel size*camera binning)/(Objective Magnification*tube lens magnification).
- Save the Macro text into a 3Dangle.txt file in the Macro subfolder in the ImageJ directory. Install the macro using Plugins>Macros>Install. Select the point selection tool (if multipoint selection tool is selected, right click to switch). Double click on the point tool icon to select the automeasure option. Alternatively, run the macro once (Plugins>Macros>3Dangle).
- Using the point selection tool set as indicated in step B3, click on the two spindle poles. Then run the macro (Plugins>Macros>3Dangle). The macro uses the first two lines of the result table to compute the angle. The calculated angle is indicated in the log window. Then, the result table is cleared.
- Acquire Z stacks with a 63x/100x PLAN APO lens. Use Nyquist/Shannon criterion for Z step calculation (0.24 μm for this lens). Image quality must be good enough so that poles are clearly identified.
- Macro
run("Point Tool...", "mark=0 auto-measure label selection=yellow");
run("Set Measurements...", " redirect=None decimal=3");
IsCalibratedImage();
x1=getResult("X", 0);
y1=getResult("Y", 0);
z1=getResult("Slice", 0);
x2=getResult("X", 1);
y2=getResult("Y", 1);
z2=getResult("Slice", 1);
xmag1=x1;
ymag1=y1;
zmag1=0;
xmag2=(x1-x2);
ymag2=(y1-y2);
zmag2=(z1-z2);
// scalar product
product = (xmag1*xmag2) + (ymag1*ymag2) + (zmag1*zmag2);
// magnitude horizontal vector 1 (points 1- to (0,0,z1)
length1 = sqrt(xmag1 * xmag1 + ymag1*ymag1 + zmag1*zmag1);
// magnitude vector 2 (points 1-2)
length2 = sqrt(xmag2 * xmag2 + ymag2*ymag2 + zmag2*zmag2);
degrees = acos(product/length1/length2);
print("3d angle is " + degrees + " degrees");
run("Select None");
run("Clear Results");
exit();
}
function IsCalibratedImage()
{
um="um";
getVoxelSize(width, height, depth, unit);
if(unit=="pixels" || unit=="microns" || unit=="micron")
{
if (width==0 || width==1)
{
Dialog.create("Image Calibration:");
Dialog.addMessage("Image has to be calibrated \nPlease enter the following parameters");
Dialog.addNumber("x , y pixel size: ", 0, 3, 5, um);
Dialog.addNumber("Z step: ", 0, 3, 5, um);
Dialog.show();
XYscale=Dialog.getNumber();
Zscale=Dialog.getNumber();
n=nSlices;
run("Properties...", "channels=1 slices=n frames=1 unit=um pixel_width="+XYscale+"
pixel_height="+XYscale+" voxel_depth="+Zscale+" frame=[0 sec] origin=0,0");
return;
}
}
}
Recipes
- 1x PBS
Made by diluting 10x PBS in MilliQ water
- 1x PBS/1% BSA
Made by addition of 1% (weight/volume) into 1x PBS
Acknowledgments
This protocol is adapted from: Bompard et al. (2013). GB was supported by a grant from ‘Fondation pour la Recherche Médicale’ (Université Montpellier 2). This work was supported by a grant MEGAPAK to NM from the ANR (Agence Nationale pour la Recherche) GENOPAT.
References
- Bompard, G., Rabeharivelo, G., Cau, J., Abrieu, A., Delsert, C. and Morin, N. (2013). P21-activated kinase 4 (PAK4) is required for metaphase spindle positioning and anchoring. Oncogene 32(7): 910-919.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Cau, J., Morin, N. and Bompard, G. (2013). Spindle Angle Measurements. Bio-protocol 3(19): e925. DOI: 10.21769/BioProtoc.925.
- Mannen, T., Yamashita, S., Tomita, K., Goshima, N. and Hirose, T. (2016). The Sam68 nuclear body is composed of two RNase-sensitive substructures joined by the adaptor HNRNPL. J Cell Biol 214(1): 45-59.
Category
Cell Biology > Cell staining > Protein
Biochemistry > Protein > Structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









