- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Enrichment of Golgi membranes from HeLa cells by sucrose gradient ultracentrifugation
Published: Vol 3, Iss 18, Sep 20, 2013 DOI: 10.21769/BioProtoc.906 Views: 15640
Reviewed by: Lin FangFanglian HeHui Zhu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Cell-free Fluorescent Intra-Golgi Retrograde Vesicle Trafficking Assay
Nathanael P. Cottam and Daniel Ungar
Nov 20, 2017 8748 Views

Cell-free Reconstitution of the Packaging of Cargo Proteins into Vesicles at the trans Golgi Network
Xiao Tang [...] Yusong Guo
Mar 5, 2020 5139 Views
Abstract
This is a protocol to extract intact Golgi Membranes from HeLa cells using sucrose gradient centrifugation. This extraction is very useful for several applications including pull-down of Golgi membrane proteins, electron microscopy and reconstitution of protein transport into an isolated system. Protocol adapted from Balch et al. (1984).
Keywords: Golgi membranesMaterials and Reagents
- HeLa cells (ATTC, Wesel, Germany)
- PBS
- 1 M Tris (pH 7.4)
- 100 mM EDTA
- Trypan Blue
- Protease inhibitor cocktail tablets (Roche, catalog number: 11836153001 )
- Breaking buffer (BB) (see Recipes)
- 29% (w/w) sucrose (see Recipes)
- 35% (w/w) sucrose (see Recipes)
- 62% (w/w) sucrose (see Recipes)
Equipment
- Cell scrapers
- Cell homogenizer (EMBL cell cracker) (EMBLEM Technology Transfer, Heidelberg)
- Cell culture microscope
- Ultracentrifuge (Beckman Coulter, model: Optima L-100K or equivalent)
- Refractometer
- SW40Ti rotor
- Centrifuge tubes
- 1 ml syringe with 20/21 G needle
Procedure
- Remove medium and wash cells 3x with PBS and 1x with Breaking buffer (BB).
- Harvest the cells by scraping and pellet the cells (for instance at 300 x g, 5 min).
- Wash pellet 2x in PBS centrifuge cells at 300 x g, 5 min.
- Wash 1x in ice-cold BB.
- Dilute the pellet 1:5 in ice-cold BB.
- Homogenize pellet with an EMBL cell cracker 20x on ice.
Note: Make sure there are no air bubbles during the homogenization. - Mix a few μl of homogenate with a trypan blue solution on a glass slide and cover it with a coverslip. Check homogenization by microscope.
Note: Plasma membrane should not be intact anymore. Cell nuclei should stain blue with Trypan Blue. There should be a lot of membrane fragments and particles in the homogenate, but the nucleus should stay intact. - Mix the homogenate with 62% sucrose
- 2 ml homogenate
- 1.83 ml of 62% ice-cold sucrose
- 41.7 μl of 100 mM EDTA (pH 7.4)
Sucrose gradients: solutions are w/w%.
Check pH of the solutions after dissolving the sucrose. - 2 ml homogenate
- Run gradient
- 4 ml homogenate (in 37% sucrose)
- 4.5 ml 35% sucrose
- 3.5 ml 29% sucrose (to the top)
- 4 ml homogenate (in 37% sucrose)
- Centrifugation: SW 40 Ti Rotor, centrifuge for 1.5 h at max speed (x g) at 4 °C.
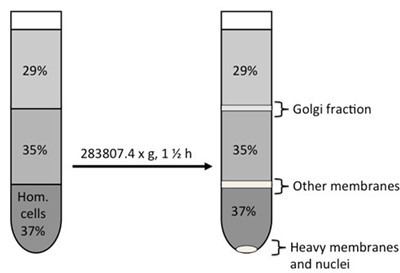
- Pull the Golgi band in 0.4 ml using a 1 ml syringe with 20/21 G needle (the Golgi band is located at the 35%/29% sucrose interphase).
- Measure protein concentration and the functional Golgi membranes can now be snap frozen in liquid N2 and stored at -80 °C.
Notes
- Avoid salts/ions in the homogenate as it may aggregate the organelles.
- Addition of high amount of sucrose affects the pH.
- Don’t homogenize too much in step 6 as organelles can break
- Proteases can leak out of the lysosomes.
- Broken organelles can reseal with other broken organelles.
- DNA can be released from nuclei which makes the sample sticky.
- Proteases can leak out of the lysosomes.
- The isolated Golgi membranes are in a buffer containing about 30% of sucrose. Therefore, if Golgi membranes need to be pelleted for further analysis, the sucrose needs to be diluted out by addition of 3 volumes of an appropriate buffer such as PBS.
Recipes
- Breaking buffer (BB)
250 mM Sucrose
10 mM Tris (pH 7.4)
Add protease inhibitor cocktail tablets - 29% (w/w) sucrose
65.08 g sucrose/200 ml
10 mM Tris (pH 7.4) - 35% (w/w) sucrose
80.60 g sucrose/200 ml
10 mM Tris (pH 7.4) - 62% (w/w) sucrose
161 g sucrose/200 ml
10 mM Tris (pH 7.4)
Note: Check all sucrose solutions with refractometer index and % of sucrose.
Acknowledgments
The protocol was adapted from the original version published by Balch et al. (1984).
References
- Balch, W. E., Dunphy, W. G., Braell, W. A. and Rothman, J. E. (1984). Reconstitution of the transport of protein between successive compartments of the Golgi measured by the coupled incorporation of N-acetylglucosamine. Cell 39(2): 405-416.
- von Blume, J., Alleaume, A.-M., Kienzle, C., Carreras-Sureda, A., Valverde, M. and Malhotra, V. (2012). Cab45 is required for Ca2+-dependent secretory cargo sorting at the trans-Golgi network. J Cell Biol 199(7): 1057-1066.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Galen, J. V. and Blume, J. V. (2013). Enrichment of Golgi membranes from HeLa cells by sucrose gradient ultracentrifugation. Bio-protocol 3(18): e906. DOI: 10.21769/BioProtoc.906.
-
von Blume, J., Alleaume, A.-M., Kienzle, C., Carreras-Sureda, A., Valverde, M. and Malhotra, V. (2012). Cab45 is required for Ca2+-dependent secretory cargo sorting at the trans-Golgi network. J Cell Biol 199(7): 1057-1066.
Category
Cell Biology > Organelle isolation > Golgi
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









