- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Colony Immunoblotting Assay for Detection of Bacterial Cell-surface or Extracellular Proteins
Published: Vol 3, Iss 17, Sep 5, 2013 DOI: 10.21769/BioProtoc.888 Views: 29262
Reviewed by: Salma HasanFanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Analysis of Direct Interaction between Viral DNA-binding Proteins by Protein Pull-down Co-immunoprecipitation Assay
Ana Lechuga [...] Modesto Redrejo-Rodríguez
Jan 5, 2018 12459 Views

Biofilm Assays on Fibrinogen-coated Silicone Catheters and 96-well Polystyrene Plates
Cristina Colomer-Winter [...] Ana L. Flores-Mireles
Mar 20, 2019 7645 Views

Construction of a Highly Diverse mRNA Library for in vitro Selection of Monobodies
Taishi Kondo [...] Hiroshi Murakami
Aug 20, 2021 3872 Views
Abstract
This simple protocol describes how to detect antigens from agar-grown bacterial colonies transferred to nitrocellulose using specific antibodies. The protocol is well suitable for detection of bacterial proteins exposed on the cell surface or secreted to the extracellular space and it can be modified also for detection of intracellular proteins. The assay can distinguish bacterial clones with different expression rates (high, medium and low) from colonies that do not express target protein. We used this assay for screening of Mat fimbriae-producing Escherichia coli mutants obtained by mini-Tn5 transposon mutagenesis and immunomagnetic separation (Lehti et al., 2013).
Keywords: Protein expressionMaterials and Reagents
- Bacteria (e.g. Escherichia coli)
- Sterile toothpicks or pipette tips
- Circular Protran BA85 nitrocellulose membrane, 0.45 μm, 82 mm in diameter (Whatman, catalog number: 10401116 )
- Albumin from bovine serum (BSA) (Sigma-Aldrich, catalog number: A7906 )
- Tween 20 (Sigma-Aldrich, catalog number: P1379 )
- Unconjugated primary antibody/antiserum (e.g. Polyclonal rabbit antiserum against Mat fimbriae of E. coli (Pouttu et al., 2001))
- Alkaline phosphatase-conjugated secondary antibody (e.g. Alkaline phosphatase-conjugated swine anti-rabbit immunoglobulins (Dako, catalog number: D0306 ))
- 1-Step BCIP/NBT solution (Pierce Antibodies, catalog number: 34042 )
- Distilled water
- Bacto Tryptone (BD Biosciences, catalog number: 211705 )
- Bacto Yeast extract (BD Biosciences, catalog number: 212750 )
- Bacto Agar (BD Biosciences, catalog number: 214010 )
- 10% (w/v) SDS solution in distilled water
- 0.5 M NaOH, 1.5 M NaCl in distilled water
- 0.5 M Tris-HCl pH 7.5, 1.5 M NaCl in distilled water
- 2x SSC (see Recipes)
- LB agar plates(see Recipes)
- Phosphate buffered saline (PBS) (see Recipes)
- Blocking buffer: 2% (w/v) BSA in PBS (see Recipes)
- Antibody dilution buffer: 1% (w/v) BSA in PBS (or the buffer as recommended in the antibody datasheet)(see Recipes)
- Washing buffer: 0.05% (v/v) Tween 20 in PBS (see Recipes)
Equipment
- 3 mm paper (Whatman 3MM, catalog number: 3030-6185 )
- Petri dishes, 90 mm (Sterilin®, catalog number: 101RT )
- Beaker, 1,000 ml (Pyrex Corning, catalog number: 1000-1L )
- Incubator (Termaks)
- Belly dancer laboratory shaker (Stovall Life Science)
- Sterile 18-gauge needle (BD Biosciences)
- Tweezers (Bochem)
Procedure
- Place identical orientation marks in three asymmetric locations on the exterior wall of two fresh agar plates (e.g. LB agar for Escherichia coli). Be sure to include orientation marks on the bottom plate (i.e. not on the lid).
- Pick each fresh colony to be tested with a sterile toothpick or pipette tip and make a short streak of about 0.5 cm in length onto the test agar plate and then onto the master agar plate. To keep the colonies in a clear physical order use a grid pattern (see Figure 1) under the plates and streak each colony in an identical position on both plates. A colony density of 100 colonies per plate (9 cm diameter) is optimal and minimizes possibility of cross-contamination between colonies. It is recommended to include a negative and positive control colony onto the plates.
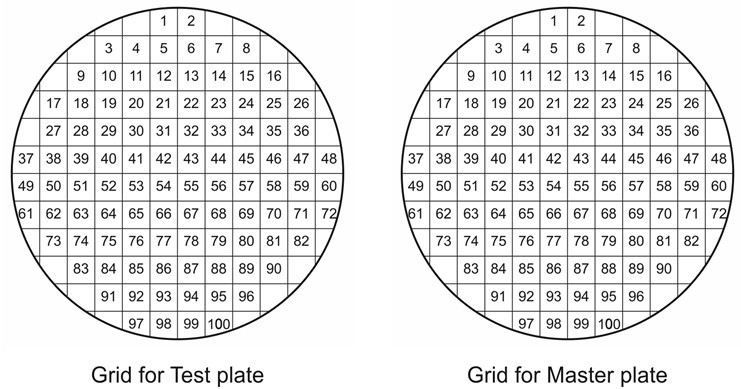
Figure 1. A grid template for precise orientation of 100 cell colonies on two agar plates. A single grid with 85 mm in diameter is used for a 90 mm Petri dish (the original picture size 19 cm x 10 cm is suitable for printing). - To grow the bacteria, invert the plates and incubate them at an appropriate temperature overnight (e.g. for Escherichia coli at 37 °C). To minimize cross-contamination among neighboring colonies, avoid overgrowth and potential formation of satellite colonies.
- Using two tweezers, place a circular nitrocellulose membrane (82 mm in diameter) carefully and evenly on top of the colonies on the test plate for 10-60 sec.
- Pre-cool the test plate at 4 °C for approximately 30 min before placing the membrane on its surface so that the agar will not adhere onto the membrane.
- To ensure contact with the colonies, take care to avoid introducing air bubbles between the membrane and the agar surface. Hold the membrane at opposite edges with two tweezers and bend it slightly, and then allow the curved membrane to make contact with the center of the plate. Finally, lower the sides gently onto the agar surface. To avoid cross-contamination and smearing of colonies, do not reorientate the membrane once it has been applied.
- Mark the membrane orientation according to the three marks on the test plate by stabbing through the membrane with a sterile 18-gauge needle.
- Save the master plate in 4 °C for later use.
- Pre-cool the test plate at 4 °C for approximately 30 min before placing the membrane on its surface so that the agar will not adhere onto the membrane.
- Using tweezers, peel off the membrane from the agar and transfer the membrane colony-side up to a beaker.
- Pour 100 ml of blocking buffer into beaker. Place the beaker on laboratory shaker (Belly dancer) and shake at speed setting 3-4 at room temperature for 5 min to remove excess of colony material.
- Decant the blocking buffer.
- Repeat steps 5a and 5b two more times.
- Pour 100 ml of blocking buffer into beaker and incubate in blocking buffer for 45 min with gently agitation on laboratory shaker (Belly dancer, speed setting 2-3) to ensure complete blocking of unoccupied protein binding sites.
- Care should be taken to prevent the membrane from drying out during incubation. Multiple membranes can be placed in a single beaker.
Note: If working with intracellular proteins, cells should be lysed before blocking. To lyse the cells, transfer the membrane colony-side up from the test plate to a Petri dish containing a sheet of 3 mm (Whatman 3 MM) paper moistened with 2-3 ml of 10% (w/v) SDS solution. Avoid introducing air bubbles. After 5 min incubation, place the membrane on top of the 3 mm paper saturated with 0.5 M NaOH, 1.5 M NaCl for 5 min. Then, neutralize by incubating two times for 5 min on the 3 mm paper soaked with 0.5 M Tris-HCl (pH 7.5), 1.5 M NaCl. Finally, place the membrane on top of the 3 mm paper saturated with 2x SSC for 15 min. Transfer the membrane colony-side up to a beaker and continue with the blocking step 5d.
- Pour 100 ml of blocking buffer into beaker. Place the beaker on laboratory shaker (Belly dancer) and shake at speed setting 3-4 at room temperature for 5 min to remove excess of colony material.
- Prepare a primary antibody dilution in antibody dilution buffer and transfer the membrane to a Petri dish containing the diluted primary antibody (one membrane per Petri dish).
- For each 82 mm-diameter membrane, use 7-10 ml of diluted primary antibody. Ensure the membrane is adequately covered with the solution to prevent it from drying out during incubation.
- The antibody dilution and incubation conditions depend on your primary antibody; please refer to primary antibody datasheet. It is recommended to start with the same conditions used for Western blotting. For optimal results, perform a titration experiment (e.g. 1:100-1:3,000) and optimize the dilution according to the results.
- A serum dilution of 1:500-1:1,000 (5-10 μl in 10 ml) and incubation for 1 hour with gently agitation at room temperature will normally be sufficient.
- For each 82 mm-diameter membrane, use 7-10 ml of diluted primary antibody. Ensure the membrane is adequately covered with the solution to prevent it from drying out during incubation.
- Wash the membrane three times in 10 ml of washing buffer for 5 min each to remove unbound antibodies.
- Prepare a secondary antibody dilution in antibody dilution buffer and replace the washing buffer with the diluted secondary antibody.
- Use 7-10 ml of diluted alkaline phosphatase-conjugated secondary antibody per 82 mm-diameter membrane.
- Please refer to secondary antibody datasheet for recommended antibody dilution buffer and recommended antibody dilution.
- Typically 1:500 to 1:2,000 dilutions of the commercial conjugates are appropriate. Incubate as in step 6.
- Use 7-10 ml of diluted alkaline phosphatase-conjugated secondary antibody per 82 mm-diameter membrane.
- Wash as in step 7.
- To visualize positive colonies reacting with the primary antibodies, replace the washing buffer with 5-10 ml of BCIP/NBT solution and incubate with gentle agitation at room temperature until the desired color develops (usually 2 to 5 min) or background colour begin to appear. The positive control should be dark purple-coloured (see Figure 2).
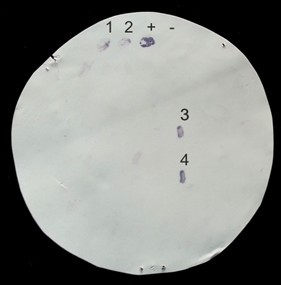
Figure 2. A representative colony immunoblotting membrane of Mat fimbriae expression in Escherichia coli MG1655-Rif derivatives. Bacteria subjected to mini-Tn5 transposon mutagenesis and enrichment with immunomagnetic particles coated with anti-Mat antibodies (available from previous work (Pouttu et al., 2001)) were grown overnight on LB agar plates at 37 °C, and colonies transferred onto a nitrocellulose membrane were left to react with anti-Mat antibodies (a dilution of 1:500) and detected with alkaline phosphatase-conjugated secondary antibodies (a dilution of 1:2,000). Colony-represents MG1655-Rif, a negative control for Mat fimbriae expression (Lehti et al., 2013). Colony + indicates IHE3034 matA(A536C) (ptet-matA), a positive control of Mat fimbriae expression (Lehti et al., 2013). Colonies 1 to 4 are candidate Mat-fimbriated MG1655-Rif Tn5 mutants. - Stop the reaction by rinsing the membrane several times in distilled water.
- Allow the membrane to air dry in the dark.
- Find the desired colonies by matching the colonies on the master plate to the reaction spots in the membrane by using orientation marks. Prepare pure cultures of the reactive colonies onto fresh agar plates.
Recipes
- 2x SSC
0.3 M NaCl, 0.03 M sodium citrate (pH 7.0) in distilled water - LB agar plates (1 L) (approximately 40-45 plates)
Add to 800 ml of H2O
10 g Tryptone
5 g Yeast extract
5 g NaCl
15 g Agar
If needed, adjust pH to 7.1 with NaOH
Add H2O to final volume of 1 L and sterilize by autoclaving (20 min, 121 °C).
Cool down to 55 °C and supplement with appropriate antibiotics.
Dispense approximately 20 ml per 90 mm-diameter Petri dish.
Store at 4 °C for up to 2 months - Phosphate-buffered saline (PBS) (pH 7.4) (1 L)
Add to 800 ml of H2O
8 g of NaCl
0.2 g of KCl
1.44 g of Na2HPO4
0.24 g of KH2PO4
Adjust the pH to 7.4 with HCl.
Add H2O to a total volume of 1 L and sterilize by autoclaving (20 min, 121 °C).
Store at room temperature - Blocking buffer
Add 10 g BSA to 500 ml of PBS and mix well.
Store at 4 °C. - Antibody dilution buffer
Add 0.2 g BSA to 20 ml of PBS and mix well. Alternatively, mix 10 ml of blocking buffer with 10 ml of PBS to make 1% BSA in PBS.
Store at 4 °C. - Washing buffer
Add 50 μl Tween 20 to 100 ml of PBS and mix well.
Store at room temperature
Acknowledgments
This protocol was modified from Sambrook, J. and Russell, D. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory). The authors would like to acknowledge University of Helsinki, Viikki Graduate School in Biosciences, the Academy of Finland (ERA-NET PathoGenoMics grant number 118982) and the European Network of Excellence in EuroPathoGenomics EPG (CEE LSHB-CT-2005-512061) for financial support.
References
- Lehti, T. A., Bauchart, P., Kukkonen, M., Dobrindt, U., Korhonen, T. K. and Westerlund-Wikstrom, B. (2013). Phylogenetic group-associated differences in regulation of the common colonization factor Mat fimbria in Escherichia coli. Mol Microbiol 87(6): 1200-1222.
- Pouttu, R., Westerlund-Wikstrom, B., Lang, H., Alsti, K., Virkola, R., Saarela, U., Siitonen, A., Kalkkinen, N. and Korhonen, T. K. (2001). matB, a common fimbrillin gene of Escherichia coli, expressed in a genetically conserved, virulent clonal group. J Bacteriol 183(16): 4727-4736.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lehti, T. A. and Westerlund-Wikström, B. (2013). Colony Immunoblotting Assay for Detection of Bacterial Cell-surface or Extracellular Proteins. Bio-protocol 3(17): e888. DOI: 10.21769/BioProtoc.888.
Category
Microbiology > Microbial biochemistry > Protein > Immunodetection
Biochemistry > Protein > Immunodetection > Immunostaining
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











