- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Retrograde and Anterograde Tracing of Neural Projections
Published: Vol 3, Iss 16, Aug 20, 2013 DOI: 10.21769/BioProtoc.870 Views: 21001
Reviewed by: Xuecai Ge

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vivo Electroporation of Skeletal Muscle Fibers in Mice
Steven J. Foltz [...] Hyojung J. Choo
Jul 5, 2023 1833 Views

Cochlear Organ Dissection, Immunostaining, and Confocal Imaging in Mice
Chenyu Chen [...] Dongdong Ren
Jan 20, 2025 3778 Views

Isolation and Imaging of Microvessels From Brain Tissue
Josephine K. Buff [...] Sophia M. Shi
Aug 5, 2025 2639 Views
Abstract
Neurons consist of four elements, the soma, dendrite, axon and terminal. They work in concert as the input (soma and dendrite) and output (axon and terminal) parts of neuronal transmission. To function and maintain neuronal activity and metabolisms, proteins and organelles should be transported from soma to terminal via anterograde axonal transport, and also from terminal to soma via retrograde transport. By utilizing these transport systems, neural projection is traced by injecting tracers into local sites of interest. Furthermore, neurochemical properties, such as glutamatergic and GABAergic, can be determined by combining retrograde and anterograde tracing with fluorescent in situ hybridization and immunofluorescence.
Keywords: In situ hybridizationMaterials and Reagents
- Chloral hydrate
- Instant glue
- Biotinylated dextran amine (BDA) (3,000 molecular weight, 10% solution in PBS) (Life Technologies, InvitrogenTM, catalog number: D-7135 )
- Alexa Fluor 488-conjugated cholera toxin subunit b (Alexa488-CTb, 0.5% solution in saline) (Life Technologies, InvitrogenTM, catalog number: C-34775 )
- Distilled H2O (dH2O)
- Pentobarbital
- Sodium azide
- Phosphate-buffered saline (PBS)
- Alexa Flour 594-conjugated streptavidin (Life Technologies, InvitrogenTM, catalog number: S-11227 )
- Normal donkey serum (10% solution in PBS) (Jackson Immuno Research, catalog number: 017-000-121 )
- Primary antibodies for immunofluorescence
Note: We almost use self-made antibodies (Kudo et al., 2012). - Secondary antibodies
Note: We use products from Invitrogen and/or Jackson Immuno Research, whose host are donkey. - Acetic anhydrate
- Triethanolamine-HCl
- Formadime
- Tris(hydroxymethyl) aminomethane (TRIS)
- Ficoll
- Polyvinylpyrrolidone
- Bovine serum albumin (BSA)
- tRNA
- Ethylenediaminetetraacetic acid (EDTA)
- N-Lauroylsarcosine sodium salt (NLS)
- Dextran sulfate
- cRNA probes (Kudo et al., 2012; Yamasaki et al., 2010)
- Sodium citrate
- Lodoamide
- DIG blocking reagent (Roche Applied Science, catalog number: 11096176001 )
- Normal sheep serum (Chemicon, catalog number: S22-100ML )
- Maleic acid
- Tyramide signal amplification (TSA) blocking reagent (PerkinElmer Life and Analytical Science, catalog number: FP1020 )
- Peroxidase-conjugated anti-DIG antibody (1:1,000 in DIG blocking buffer) (Roche Diagnostics, catalog number: 11207733910 )
- Cyanine 3 (Cy3) Amplification Reagent (PerkinElmer Life and Analytical Science, catalog number: FP1170 )
- 1x Plus Amplification Diluent (PerkinElmer Life and Analytical Science, catalog number: FP1135 )
- H2O2
- Anti-Alexa Fluor 488 antibody for detection of Alexa488-CTb (raised in rabbit) (1:1,000 dilution) (Invitrogen, catalog number: A-11094 )
- TOTO-3 iodide (642/660) (1:50 in PBS) (Life Technologies, InvitrogenTM, catalog number: T3604 )
- Primary antibody solution (1 μg/ml in PBS-T)
- Fluorophore-linked secondary antibody solution (1:200 in PBS-T)
- Digoxigenin (DIG)-labeled cRNA probes
- Dimethyl sulfoxide (DMSO)
- Blocking reagent (Roche Applied Science, catalog number: 11096176001)
- Saline (see Recipes)
- 3.5% chloral hydrate (see Recipes)
- 0.1 M PB (see Recipes)
- 5 N NaOH (see Recipes)
- 4% Paraformaldehyde (PFA) (see Recipes)
- 20% Tween-20 (see Recipes)
- PBS containing 0.1% Tween-20 (PBS-T) (see Recipes)
- 0.25% acetic anhydrate in 0.1 M triethanolamine-HCl (pH 8.0) (see Recipes)
- 1 M Tris-HCl (pH 7.4 or 8.0) (see Recipes)
- 5 M NaCl (see Recipes)
- 0.5 M ethylenediaminetetraacetic acid (EDTA) (see Recipes)
- Hybridization buffer (see Recipes)
- Standard saline citrate (SSC) (see Recipes)
- NaCl-Tris-EDTA (NTE) buffer (see Recipes)
- Tris-NaCl-Tween (TNT) buffer (see Recipes)
- 20 mM iodoamide in NTE buffer (see Recipes)
- DIG blocking solution (see Recipes)
- 0.5% TSA blocking buffer (see Recipes)
- Cy3-TSA amplification solution (see Recipes)
Equipment
- Scissors
- Surgical knife
Note: We use set of stainless-steel mess handle (Feather, No.3) and stainless-steel spare blade (Feather, No.14). - Syringe
- Syringe needle
- Stereotaxic instrument (Narishige, model: SR-5M )
- Surgical cotton
- Pneumatic pump (Pneumatic Picopump) (World Precision Instruments, model: PV800 )
- Puller (Narishige, model: PC-10 )
- Glass pipette (Narishige, model: G-1.2 )
- Peristaltic pump (ATTO Corporation, model: SJ-1211H )
- Cork board
- Microslicer (Leica Microsystems, model: VT1000S )
- Razor blade
- 10 ml tube
- Confocal laser-scanning microscope (Olympus, model: FV1000 )
- Hybridization oven (Bellco, model: 7930-00110 )
- Water bath
- Fluorescence microscope
Note: Fluorescence signals of Alexa Fluor 488 and Cy3 or Alexa Fluor 594 are observed through fluorescence mirror units (Olympus, model: U-MWIBA3 , U-MWIG3 ). - Aspirator
Software
- Confocal software (Olympus, FV10-ASW, ver.1.7)
Procedure
- Injection of anterograde or retrograde tracer
- Prepare the tracer solution and all equipment needed (scissors, surgical knife, syringe needles etc).
Note: BDA and Alexa488-CTb are used as an anterograde or retrograde tracer, respectively. Spin down tracer solutions before use. - Anesthetize an animal with 3.5% chloral hydrate (350 mg/kg body weight, i.p.).
- Check the animal being anesthetized by loss of righting reflex and lack of response to hitching its paws.
- Place the anesthetized animal on stereotaxic instrument with locking the nose and ears.
Note: First, insert the auxiliary ear bar into the mouse ear canal and fix it tightly, but be careful not to insert it too deeply to injure the inner ear. Next, align the vertical level of the nose clamp and hook the mouse tooth on it, and then hold down the nose. Check whether the mouse head do not move wobbly to finish fixation. - Shave the hair and incise the scalp along the rostro-caudal axis.
- Clip the incised scalp using syringe needles bent into a hook and remove lamina.
Note: To remove lamina, snick it by scissors and wipe the surface of the skull bone by surgical cottons. - Using a surgical knife cut the skull bone and make a square hole at the position of tracer injection.
- Prepare a glass pipette using a puller.
- Fill a glass pipette with ~1 μl of the tracer solution by a capillary phenomenon and attach it to a tube connected with a pneumatic pump.
Note: If the tip of the pipette is too fine to draw the tracer solution by a capillary phenomenon, snap the tip slightly. Moreover, too fine tip often prevents smooth injection as tiny concomitants get jammed in the pipette. - Position the tip of the glass pipette on the bregma and then stereotaxically insert the pipette into the target region.
Note: To make injection space, insert the pipette 0.1 mm deeper and turn back to the target region. - Inject the tracer solution by air pressure at 10 psi with 5 sec intervals for 1 min.
- Leave the pipette inserted for 15 min, and then pull out it carefully.
- Replace the removed skull bone on the square hole and suture the scalp.
Note: Instant glue is used for closing incision site as substitute for a surgical suture. - Release and place the animal back into a home cage.
- Keep the animal for several days until fixation.
Note: Determine the period of survival depending on the distance of projection.
In our experiments (between the bed nucleus of the stria terminalis (BST) and the ventral tegmental area (VTA)), BDA-injected and Alexa488-CTb-injected animals are incubated for at least 4 and 2 days, respectively (Kudo et al., 2012).
- Prepare the tracer solution and all equipment needed (scissors, surgical knife, syringe needles etc).
- Perfusion and section preparation
- Place a beaker containing 4% PFA fixative solution (100 ml for each mouse) in ice bath. Set on peristaltic pump a silicone tube with one end put into the fixative beaker and the other end equipped with a 25 G syringe needle.
- Prepare all equipment needed (scissors, forceps, syringe needles to impale hands/feet etc).
- Run the peristaltic pump and fill up tubes and the syringe needle with the fixative solution.
- Deeply anesthetize the animal with overdosed pentobarbital (100 mg/kg of body weight, i.p.).
- After confirming the animal asleep, impale the animal’s hands and feet on a cork board with syringe needles.
- Open the abdominal cavity by horizontal cutting of the belly skin and muscles. Then cut the skin of the chest along the midline up to the jaw, and detach the skin from the chest wall.
- Cut the diaphragm to open the thoracic cavity, and then cut both sides of the chest wall to expose the heart. It is important not to injure the internal thoracic artery, which runs vertically along the sternum.
- Snick the wall of the right auricle by sharp scissors or the pit of syringe needle, and prick the left ventricle with a syringe needle connecting to the fixative beaker.
- Start transcardial perfusion for 10 min, so that 3 fixative volumes of the body weight run in 10 min.
- Excise a fixed brain and post-fix it for 2 h. Using a razor blade, divide the brain into two blocks with one containing a tracer-injected site and another containing neural regions of interest.
Note: In our experiments, we divided the brain into two parts containing the BST and VTA, respectively, by coronal cutting between the hypothalamus and mammillary body (Kudo et al., 2012). - Prepare sections of the fixed brain (50 μm in thickness) using a microslicer, whose buffer bath is filled with 0.1 M PB.
Note: Especially in free-floating in situ hybridization experiment, the fixed brain block and sections should be kept in PB. When PBS is used for buffer, detection sensitivity of mRNA signal tends to be lowered. - Collect sections in 24-well plate and store in 0.1 M PB containing 0.1% sodium azide.
- Place a beaker containing 4% PFA fixative solution (100 ml for each mouse) in ice bath. Set on peristaltic pump a silicone tube with one end put into the fixative beaker and the other end equipped with a 25 G syringe needle.
- Anterograde tracing combined with fluorescence immunohistochemistry
Note: This method is used to determine neurochemical properties of axon terminals, which are projected from anterograde tracer-injected regions. Vesicular glutamate transporters (VGluTs) and vesicular inhibitory transporter (VIAAT) are frequently used as excitatory (glutamatergic) and inhibitory (GABAergic and glycinergic) terminal markers, respectively (Kudo et al., 2012; Figure 1 in this manual). By using other neurochemical markers for combination with anterograde tracing, one can also determine whether the projection is cholinergic, serotonergic, dopaminergic, adrenergic, histeminergic, or peptidergic.- Prepare sections from BDA-injected brains.
- Select sections containing the injected region and visualize BDA signals by incubation in Alexa Flour 594-conjugated streptavidin (1:500 in PBS) for 10 min. Photograph the injection site of BDA using a fluorescence microscope.
- Submerge 1~3 sections for neurochemical testing in a 10 ml tube containing PBS containing 0.1% Tween-20 (PBS-T) for 10 min.
- Block with 10% normal donkey serum in PBS for 20 min.
Note: Blocking and antibody solutions are ~0.5 ml per tube, and washing solutions are ~10 ml per tube in each wash. Solution exchange is efficient by using aspirator, but should be carefully done not to aspirate or dry up sections. - Incubate in primary antibody solution (1 μg/ml in PBS-T) overnight.
- Wash with PBS-T for 5 min three times.
- Incubate in fluorophore-linked secondary antibody solution (1:200 in PBS-T) for 2 h for neurochemical marker detection.
- Wash with PBS-T for 5 min three times.
- Wash with PBS briefly.
- Incubate in Alexa Flour 594-conjugated streptavidin (1:500 in PBS) for 10 min for BSA detection.
- Wash with PBS for 5 min three times.
- Mount sections on glass slides, make coverslip, and observe using a fluorescence microscope.
Note: Images are captured using a confocal laser scanning microscope, digitized at 12 bit resolution into an array of 640 x 640 pixels (pixel size, 0.1 μm). To investigate the neurochemical characteristic of BDA-labeled axon terminals, take images (x60 magnifications, x3 zoom) of the traced region from each slice. For analysis, we counted terminal marker-positive boutons whose center point matched with that of BDA labeling. Bouton-like structures which are only labeled with BDA are often observed. If they are not labeled with other terminal markers, they should not be used for counting (Figure 1).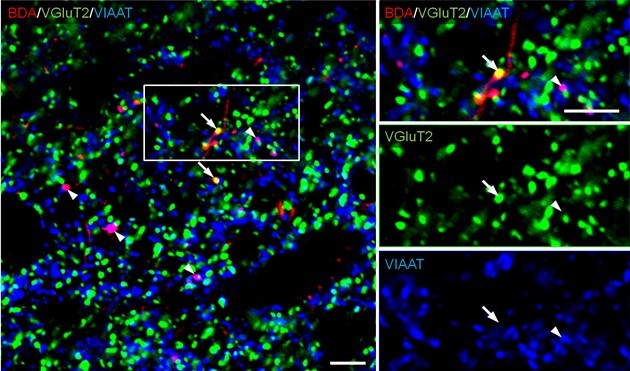
Figure 1. Combined anterograde tracer labeling and immunofluorescence. BDA is injected into the bed nucleus of the stria terminalis (BST) and anterogradely transported to the ventral tegmental area (VTA). In sections containing the VTA, BDA staining (red) and immunofluorescence for type 2 vesicular glutamate transporter (VGluT2; green) and vesicular inhibitory amino acid transporter (VIAAT; blue) are performed. Images are used for counting (x60 magnifications, x3 zoom). VGluT2-positive BDA axon terminals (arrows) and VIAAT-positive BDA axon terminals (arrowheads) are identified as glutamatergic and GABAergic afferents from the BST, respectively. Scale bars: 10 μm.
- Prepare sections from BDA-injected brains.
- Retrograde tracing combined with free-floating fluorescent in situ hybridization (FISH)
Note: This method is used to determine neurochemical properties of neurons, which project their axons to retrograde tracer-injected regions. It is recommended to prepare sections and start FISH incubation on the same day, because detection sensitivity is drastically decreased after sectioning. Expression of VGluTs and glutamic acid decarboxylase (GAD) (or VIAAT) mRNAs are used to determine the neurochemical properties of excitatory and inhibitory neurons, respectively (Kudo et al., 2012; Figure 2 in this paper).- Prepare sections from Alexa488-CTb-injected brains.
- Check the injection site of Alexa488-CTb using a fluorescence microscope.
- Submerge a single section in a 10 ml tube containing PB.
Note: It is recommended that one tube is used for one section, because more than two sections per tube decrease reaction sensitivity. For free-floating FISH, solutions for hybridization, antibody reaction, inactivation of peroxidases, fluorescence detection and counterstaining are ~0.5 ml per tube, and washing solutions are ~10 ml per tube in each wash. Solution exchange is efficient by using aspirator, but should be carefully done not to aspirate or dry up sections. - Acetylate sections with 0.25% acetic anhydrate in 0.1 M triethanolamine-HCl (pH 8.0) for 10 min.
Note: Prepare the solution at time of use. - Prehybridize in hybridization buffer for 1 h.
- Hybridize in hybridization buffer supplemented with digoxigenin (DIG)-labeled cRNA probes at a dilution of 1:1,000, performed at 63.5 °C for 12 h.
Note: Preparation of DIG-labeled cRNA probes is described in previous our reports (Kudo et al., 2012; Yamasaki et al., 2010). Hybridization is performed in a hybridization oven. Cover the tube tip with parafilm to avoid drying off. - Wash with 5x SSC for 30 min, 4x SSC containing 50% formamide (Formamide 1) for 40 min, 2x SSC containing 50% formamide (Formamide 2) for 40 min and 0.1x SSC for 15 min, performed at 61 °C.
Note: Solutions are prewarmed at 61 °C in hot bath, and washing steps are performed in hot bath. - Incubate in 0.1x SSC for 15 min, NTE buffer for 20 min, 20 mM iodoamide in NTE buffer for 20 min, NTE buffer for 10 min and TNT buffer for 10 min, performed at room temperature (RT).
Note: All subsequent steps are performed at RT. - Block sections with DIG blocking buffer for 30 min and 0.5% TSA blocking buffer for 30 min.
- Incubate in peroxidase-conjugated anti-DIG antibody in DIG blocking buffer (1:1,000) for 2 h.
- Wash with TNT buffer for 15 min twice.
- Detect signals for peroxidase using Cy3-TSA plus amplification kit system (see Recipe 20) for 10 min, performed in shade.
- Wash with TNT buffer for 5 min three times.
- For inactivation of residual peroxidase activities, incubate in 3% H2O2 in TNT buffer for 30 min.
Note: This inactivation step is important to enhance contrast of signals. If this step is skipped, background signals remain strong as well as true-positive signals. - Wash with TNT buffer for 5 min three times.
- Check the fluorescence signals for target mRNAs using a fluorescence microscope.
Note: Because the fluorescence of Alexa488-CTb becomes weak and readily extincts after hybridization and the post-hybridization wash, the tracer should be detected by immunofluorescence using anti-Alexa Fluor 488 antibody, as shown in Procedure III. - After immunofluorescence for Alexa488-CTb, counterstain with TOTO-3 (1:50 in PBS) for 20 min.
- Mount sections on glass slides and observe using a fluorescence microscope.
Note: To investigate the neurochemical composition of retrogradely traced neurons, collect tiled images to cover the whole traced region (x20 magnifications, x1.3 zoom), and counted the number of positive cells having the nucleus (show TOTO-3 signal) (Figure 2).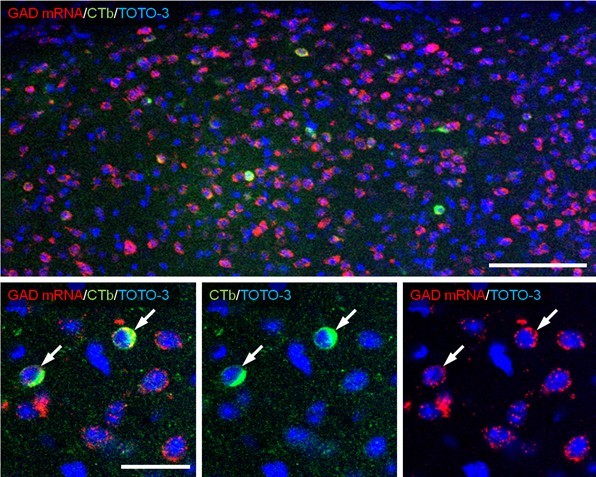
Figure 2. Combined retrograde tracer labeling and fluorescence in situ hybridization. Alexa488-CTb is injected into the VTA and retrogradely transported to the BST. In sections containing the BST, immunofluorescence for Alexa Fluor 488 (green), fluorescence in situ hybridization for GAD mRNA (red) and counterstaining by TOTO-3 (blue) are performed. The top image is a part of tiled image used for counting (x20 magnifications, x1.3 zoom) and bottom images are higher magnification (x60 magnifications, x1.0 zoom). Neurons co-labeled for GAD mRNA and CTb (arrows) are identified as VTA-projecting GABAergic neurons. Scale bar: top, 100 μm; bottom, 30 μm.
- Prepare sections from Alexa488-CTb-injected brains.
Recipes
- Saline
NaCl 9 g/L of dH2O - 3.5% chloral hydrate
Chloral hydrate 1.75 g/50 ml of dH2O - 0.1 M PB
To make 1 L of 0.1 M PB
Mix 2.95 g of NaH2PO4.2H2O and 29 g of Na2HPO4.12H2O
Add dH2O to 1 L and stir ~ 30 min
Store at 4 °C - 5 N NaOH
NaOH 40 g/200 ml of dH2O - 4% PFA
To make 1 L of PFA
Make 500 ml of 8% PFA (Solution (a))
Heat 500 ml dH2O to 80 °C (Do NOT boil)
Add 40 g of PFA powder and stir ~10 min
Add 250 μl of 5 N NaOH and keep stirring until the solution gets clear
Next, make 500 ml of 0.2 M PB (Solution (b)) in another beaker
Mix 2.95 g of NaH2PO4.2H2O and 29 g of Na2HPO4.12H2O
Add dH2O to 500 ml and stir ~ 30 min
Mix solution (a) and solution (b) and stir ~5 min
Filtrate and store at 4 °C - PBS
To make 1 L of 10x PBS stock solution
Mix 87 g of NaCl, 3.1 g of NaH2PO4.2H2O and 28.7 g of Na2HPO4.12H2O
Add dH2O to1 L and stir ~ 2 h
Store at RT
Dilute this 10x stock solution by 1/10 using dH2O - 20% Tween-20
Mix 20 ml of Tween-20 with 80 ml of ddH2O by stirring
Store at 4 °C - PBS containing 0.1% Tween-20 (PBS-T)
Add 2.5 ml of 20% Tween-20 with 500 ml of PBS
Store at RT - 0.25% acetic anhydrate in 0.1 M triethanolamine-HCl (pH 8.0)
To make 100 ml of acetylation solution
Mix triethanolamine-HCl with 100 ml of dH2O by stirring
Add 950 μl of 5 N NaOH and make sure that pH is 8.0
Add 250 μl of acetic anhydride (*add just before use) and stir ~3 min - 1 M Tris-HCl (pH 7.4 or 8.0)
To make 1 L of Tris-HCl buffer
Mix 121.1 g of Tris base (tris(hydroxymethyl)aminomethane) with 800 ml of dH2O
pH to 7.4 or 8.0 with HCl
Add dH2O to 1 L
Autoclave and then store at RT - 5 M NaCl
To make 1 L of 5 M NaCl
Mix 292.2 g of NaCl with 800 ml of dH2O by stirring
Add dH2O to 1 L and stir until they dissolve completely
Autoclave and then store at RT - 0.5 M ethylenediaminetetraacetic acid (EDTA)
To make 500 ml of 0.5 M EDTA
Mix 93.1 g of EDTA with 400 ml of dH2O
pH to 8.0 with 5 N NaOH
Add dH2O to 500 ml
Autoclave and then store at RT - Hybridization buffer
To make 500 ml of hybridization buffer
Autoclave two 500 ml beakers; one is filled with 250 ml of dH2O and another is empty
After beakers cool down, pour 80 ml of autoclaved dH2O and 250 ml of formamide into the empty beaker
Add reagents described on the following table
*100x Denhardt'sReagentQuantity 1 M Tris-HCl (pH 8.0) 16.5 ml tRNA 100 mg 100x Denhardt's* 5 ml 5M NaCl 60 ml 0.5 M EDTA 1 ml NLS 0.5 g Dextran sulfate 50 g
Add autoclaved dH2O to 500 ml and then dissolve on shaking for ~24 h.Reagent Quantity (for 50 ml) Final concentration (100x) Ficoll 1 g 2% (w/v) polivinylpyrrolidone 1 g 2% (w/v) BSA 1 g 2% (w/v) dH2O to 50 ml
Filtrate and then store at -30 °C. - Standard saline citrate (SSC)
To make 1 L of 20x SSC stock solution
Mix 175.3 g of NaCl and 88.2 g of sodium citrate
Add dH2O to1 L and stir ~ 2 h
Autoclaved and then store at RT
Dilute this 20x stock solution as described on the following table
Store at RT5x SSC
(for 1 L)0.1x SSC
(for 1 L)Formamide 1
(for 150 ml)Formamide 2
(for 150 ml)20x SSC 250 ml 5 ml 30 ml 15 ml 20% Tween-20 25 μl 25 μl 7.5 μl 7.5 μl formamide -- -- 75 ml 75 ml dH2O to 1 L to 1 L to 150 ml to 150 ml - NaCl-Tris-EDTA (NTE) buffer
To make 1 L of NTE buffer
Mix 100 ml of 5 M NaCl, 10 ml of 1 M Tris-HCl (pH 8.0), 10 ml of 0.5 M EDTA and 25 μl of 20% Tween-20
Add dH2O to1 L and store at RT - Tris-NaCl-Tween (TNT) buffer
To make 1 L of TNT buffer
Mix 30 ml of 5 M NaCl, 100 ml of 1 M Tris-HCl (pH 7.4) and 25 μl of 20% Tween-20
Add dH2O to1 L and store at RT - 20 mM iodoamide in NTE buffer
Iodoacetamide 0.37 g/100 ml of NTE buffer - DIG blocking solution
Normal sheep serum: 10% blocking reagent*: TNT buffer = 1:1:8
*10% blocking reagent (for 10 ml)- Prepare maleic acid buffer
To make 500 ml of maleic acid buffer
Mix 58.1 g of maleic acid and 43.9 g of NaCl with 400 ml of dH2O
pH to 7.5 with 5 N NaOH - Dissolve 1 g of blocking reagent to 10 ml with the maleic acid buffer with shaking and heating
- Autoclave and then store at -80 °C
- Prepare maleic acid buffer
- 0.5% TSA blocking solution
Add 0.5 g of blocking reagent to 100 ml of TNT buffer
To dissolve the blocking reagent, heat to 60 °C for 1 h with stirring
Store at -20 °C - Cy3-TSA amplification solution
Add 60 μl of DMSO to Cy3 Amplification Reagent (Cy3 solution; store at 4 °C)
Dilute this Cy3 solution in 1x Plus Amplification Diluent (1:200)
Acknowledgments
This protocol is adapted from Kudo et al. (2012).
References
- Kudo, T., Uchigashima, M., Miyazaki, T., Konno, K., Yamasaki, M., Yanagawa, Y., Minami, M. and Watanabe, M. (2012). Three types of neurochemical projection from the bed nucleus of the stria terminalis to the ventral tegmental area in adult mice. J Neurosci 32(50): 18035-18046.
- Yamasaki, M., Matsui, M. and Watanabe, M. (2010). Preferential localization of muscarinic M1 receptor on dendritic shaft and spine of cortical pyramidal cells and its anatomical evidence for volume transmission. J Neurosci 30(12): 4408-4418.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Kudo, T. and Watanabe, M. (2013). Retrograde and Anterograde Tracing of Neural Projections. Bio-protocol 3(16): e870. DOI: 10.21769/BioProtoc.870.
- Kudo, T., Uchigashima, M., Miyazaki, T., Konno, K., Yamasaki, M., Yanagawa, Y., Minami, M. and Watanabe, M. (2012). Three types of neurochemical projection from the bed nucleus of the stria terminalis to the ventral tegmental area in adult mice. J Neurosci 32(50): 18035-18046.
Category
Neuroscience > Neuroanatomy and circuitry > Animal model
Cell Biology > Tissue analysis > Tissue isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link