- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Maize Embryo Transient Transformation by Particle Bombardment
Published: Vol 3, Iss 16, Aug 20, 2013 DOI: 10.21769/BioProtoc.865 Views: 10207
Reviewed by: Tie Liu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Cellulase and Macerozyme-PEG-mediated Transformation of Moss Protoplasts
Rituraj Batth [...] Henrik Toft Simonsen
Jan 20, 2021 4543 Views

A Fast and Easy Method to Study Ralstonia solanacearum Virulence upon Transient Gene Expression or Gene Silencing in Nicotiana benthamiana Leaves
Wenjia Yu and Alberto P. Macho
Aug 5, 2021 4700 Views

Agrobacterium-mediated Genetic Transformation of Cotton and Regeneration via Somatic Embryogenesis
Alka Srivastava [...] Praveen C. Verma
May 20, 2023 4358 Views
Abstract
Particle bombardment has been shown to be a useful method to study gene promoter regulatory elements by transient transformation of maize embryos with different constructions of gene promoters fused to a gene reporter. DNA to transfer is coated to high density gold microparticles and introduced into cells when accelerated by a helium pulse. This method allows a first rapid approach, avoiding time consuming stable transformation of maize plants and also allows quantitative promoter expression analysis by a histochemical or fluorometric assay.
Keywords: Particle bombardmentMaterials and Reagents
- Plant Material: Maize plants from W64A pure inbred line
- Ethanol absolute, gradient HPLC grade (Scharlau, S.A. UN1170)
- 1.0 μm Gold Microcarriers (Bio-Rad Laboratories, catalog number: 165-2263 )
- Rupture Disks 900 psi (Bio-Rad Laboratories, catalog number: 165-2328 )
- Macrocarriers (Bio-Rad Laboratories, catalog number: 165-2335 )
- Stopping Screens (Bio-Rad Laboratories, catalog number: 165-2336 )
- Murashige and Skoog medium (M&S) with vitamins (Duchefa, catalog number: M0222 )
- Spermidine free base (Sigma-Aldrich, catalog number: S-2626 )
- Sodium hypochlorite, solution 5-9% Cl active (Carlo Erba, catalog number: CH0223 )
- Cell culture dishes 60 mm x 15 mm Style treated polystyrene (Corning Inc., catalog number: 430166 )
- X-GlcA sodium trihydrate (Duchefa, catalog number: X-1406 )
- X-glucuronide: 5 bromo-4-chloro-3-indolyl-β-D-glucoronic sodium salt 3H2O
- N,N-dimethylformamid for spectroscopy (Merck KGaA, catalog number: 102937 )
- MUG: 4 methyl-umbelliferyl-B-D-glucuronide hydrate C (Sigma-Aldrich, catalog number: M9130 )
- Potassium hexacyanoferrate (III) (Merck KGaA, catalog number: 104973 )
- Potassium hexacyanoferrate (II) trihydrate (Merck KGaA, catalog number: 104984 )
- 4-Methylumbelliferone sodium salt (4-MU) (Sigma-Aldrich, catalog number: M-1508 )
- Luciferase Assay Reagent (Promega Corporation, catalog number: E1500 )
- CDTA: trans-1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid (Sigma-Aldrich, catalog number: D0922 )
- Protein Assay Dye Reagent Concentrate (Bio-Rad Laboratories, catalog number: 500-0006 )
- Histochemical analysis detection buffer (see Recipes)
- Fluorometric lysis buffer (see Recipes)
- Fluorometric analysis reaction buffer (see Recipes)
- Fluorimetric analysis stopping buffer (see Recipes)
- MSO medium (see Recipes)
- 1x Luciferase lysis buffer (see Recipes)
Equipment
- BIORAD PDS-1000/He system (Biolistic Delivery Systems)
- Laminar Flow Cabinet Telstar AH-100
- Vacuum pump Telstar 50/60 Hz
- Cylinder compressed helium gas (UN1046) 117 Kg (LYNDE)
- RAYPA Bathwater sonicator 50 W
- MINI-SECOEUR IKA MS2 minishaker with a support that allows simultaneous agitation of different Eppendorf tubs
- MiniSpin eppendorf centrifuge
- ND-1000 Spectrophotometer (NanoDrop)
- Spectra Max M3 apparatus (bioNova cientifica S.L.)
- Olimpus Stereomicroscope SZX16
- Screw cylinder polypropylene microtube with attached O-Ring cap and conical base (Sarstedt, catalog number: 72.693.105 )
Procedure
- Embryo excision from the maize kernel
- Submerge and wash maize ears 16-18 days after manual pollination (dap) successively in different solutions as follows at room temperature:
- 1 min with ethanol 96%. Discard.
- 10 min with 25% Sodium hypochlorite. Discard.
- 5 min sterile water (x 3 times). Discard.
- 1 min with ethanol 96%. Discard.
- Detach grains with gloved hands from the washed ears and embryos from the grains with the help of a sterile cutter in a sterile Petri glass dish. Place 9 embryos/dish in a 3 x 3 array in the center of cell culture dishes (60 mm diameter) containing MSO medium. The embryo axis side in contact with the medium. Manipulations are done under a Laminar Flow Cabinet.
- Maintain plates at 21-23 °C for 24 h in the dark before bombardment to allow embryos to be recovered of excision treatment. The described conditions have been shown not to alter the embryogenesis programme of the studied maize embryos (Jose-Estanyol et al., 2012).
- Submerge and wash maize ears 16-18 days after manual pollination (dap) successively in different solutions as follows at room temperature:
- Microcarrier stock preparation (at room temperature)
- Add 1 ml ethanol (HPLC) to 60 mg of Gold Microcarriers in a cylinder microtube with attached ring cap and a conic base. Vortex at 2,200 rpm with the minishaker for 10 min. Avoid the use of hydrated ethanols.
- Centrifuge in the MiniSpin Eppendorf Centrifuge for 1 min at 10,000 rpm.
- Remove the Ethanol with a pipette, without removing microcarriers sediment. Discard ethanol.
- Add 1 ml sterile water. Vortex 1 min. Sediment by centrifugation as in step 2-b. Discard water (Repeat 3 times).
- Suspend microcarriers in 1 ml of water. Vortex 1 min. Make aliquots (30 μl) in conical tubes from the homogeneous suspension.
- Store at -20 °C.
- Add 1 ml ethanol (HPLC) to 60 mg of Gold Microcarriers in a cylinder microtube with attached ring cap and a conic base. Vortex at 2,200 rpm with the minishaker for 10 min. Avoid the use of hydrated ethanols.
- Microcarriers coating with the plasmid DNA of interest
- Quantify DNA stocks (prepared by using a commercial plasmid prep kit) of plasmids sharing the studied promoters fused to a gene reporter, usually the beta-glucoronidase enzyme, with a nanodrop Spectrophotometer.
- Sonicate defreezed microcarrier aliquots for 3 min in a bathwater sonicator.
- Add succesivelly to the microcarrier aliquots:
12.5 μl DNA in TE buffer (1 μg/μl)
95 μl of water
125 μl CaCl2 2.5 M (while vortexing: Open the tube and decrease speed to avoid sample lost while you add solutions)
25 μl spermidine 0.1 M (while vortexing) - Vortex for 3-5 min.
- Allow sedimentation on ice to minimize ethanol evaporation for 15-20 min.
- Discard solution without disturbing microcarrier sediment as in step 2-c.
- Add 500 μl ethanol (HPLC). Vortex 10-20 sec. Allow sedimentation during 15-20 min on ice.
- Discard ethanol as in step 2-c.
- Repeat with 200 μl ethanol (HPLC).
- Discard ethanol as in step 2-c.
- Finally add 40 μl ethanol (HPLC).
- Sonicate in the bathwater sonicator 3 sec (Repeat 3 times).
- Vortex 3 min.
- Distribute the 40 μl homogenous solution of DNA coated microcarriers in ethanol (HPLC) (step 3-j) between the surface center of three macrocarriers (≈ 10 μl/macrocarrier) on a flat leveled surface. Macrocarriers have been washed in 70% ethanol and dried, beforehand. Let dry. This will allow the bombardment of three different samples (three cell culture dishes, each with 9 embryos in MSO medium).
- When microcarriers have dried on macrocarriers surface (5-10 min), they are introduced inside previously autoclaved macrocarriers holders and are ready to be used for maize embryo bombardment.
- Quantify DNA stocks (prepared by using a commercial plasmid prep kit) of plasmids sharing the studied promoters fused to a gene reporter, usually the beta-glucoronidase enzyme, with a nanodrop Spectrophotometer.
- Maize embryos 16-18 dap particle bombardment
- Wash stopping screens and rupture disks with 70% ethanol. Locate in the adaptor the stopping screen and the macrocarrier holder with microcarriers attached to the macrocarrier. Locate the rupture disk at the end of the acceleration tube.
- Proceed to the bombardement of the cell culture dishes with the maize embryos in MSO medium with the microcarriers coated with the different studied plasmids following PDS-100/He System instructions (http://www.Bio-Rad.com/biolostics).
- Bombardement parameters are as follows:
Gap distance 1.0 cm Macrocarrier travel distance 1.5 cm Target distance 9.5 cm Gold microcarriers 1.0 μm Camera partial vacuum 0.1 atm Rupture Disks 900 psi Helium pressure at the regulator 1,100 psi - After bombardment and before the quantitative fluorometric or histochemical analysis, Petri dishes are incubated in the dark at 21-23 °C for 24 h to allow expression of the reporter gene. Fluorometric assay is very useful for strong promoters but for low or medium ones, the histochemical analysis can be more sensitive and avoid dilution of the signal (present mainly in the scutellum first cell layer of the bombarded embryos, penetration depth). In histochemical analysis better quantification can be achieved when both the number and the diameter of blue spots (see Figure 1) are quantified (for example: twelve basic units for spots with 80 μm diameter, six basic units for spots with 40 μm diameter, 3 basic units for spots with 20 μm diameter and one basic unit for spots with less than 20 μm diameter).
- The validity of results is evaluated by the reproducibility of the results. This is achieved by the mean value of different experiments (3 to 4) and their standard deviation.
- Finally significant differences between constructs or conditions (p-value) are calculated from the means of different experiments in a Student’st test (http://www.physics.csbsju.edu).
- Different efficiency has been observed in different maize varieties. Efficiency has shown to be in an inverse rapport with the scutellar hydration level degree of the different maize varieties during embryo development.
- Wash stopping screens and rupture disks with 70% ethanol. Locate in the adaptor the stopping screen and the macrocarrier holder with microcarriers attached to the macrocarrier. Locate the rupture disk at the end of the acceleration tube.
- Quantitative histochemical analysis
- Embryos from each bombarded dish are transferred to 2 ml eppendorf tubes with 1 ml of histochemical analysis detection buffer and incubated overnight at 37 °C.
- Blue spots of different intensity appear as result of the precipitation of product derivates reaction, see Figures 1-2 (This reaction is greatly enhanced by using an oxidation catalyst such as a potassium ferricyanide/ferrocyanide mixture).
- To avoid diffusion of the blue spots on the surface of the embryos, they are transferred to a 50% glycerol solution in water. Blue spots are quantified by observation with an OLYMPUS Stereomicroscope SZX16. Stained embryos are stored at 4 °C.
- As control one set of embryos can be bombarded with a constitutive promoter for monocots, such as OsActine: GUS-nos ter, to easily evaluate the efficiency of the microcarrier batch and of each specific experiment.
- Histochemical analysis examples:
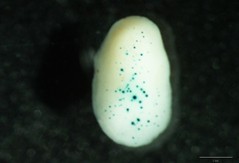
Figure 1. Maize embryos bombarded with a promoter with a medium-low expression level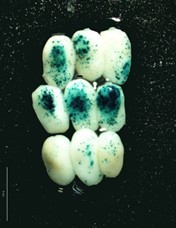
Figure 2. Set of maize embryos bombarded with OsActine::Gus-nos terconstitutive strong promoter
- Embryos from each bombarded dish are transferred to 2 ml eppendorf tubes with 1 ml of histochemical analysis detection buffer and incubated overnight at 37 °C.
- Quantitative fluorometric analysis
- Embryos from each bombarded dish are frozen in liquid N2 and stored at -80 °C until analysis.
- The nine frozen embryos from a bombarded dish are grinded in 300 μl fluorometric lysis buffer.
- Centrifuge 15 min 13,000 rpm in the MiniSpin Eppendorf Centrifuge at 4 °C.
- Freeze the clear extract solution at -80 °C in aliquots of 40 μl, until beta-glucoronidase quantification. Sediment is rejected.
- For fluorometric analysis 20 μl of clean extract are added to 80 μl fluorometric analysis reaction buffer and warmed at 37 °C.
- Aliquots of 20 μl are taken at different time courses (10 (zero), 60, 180, 360 min).
- Reaction is stopped by an addition of 180 μl of fluorimetric analysis stopping buffer to each taken aliquot and stored 4 °C until analysis.
- Samples are transferred to a microtiter plate to measure fluorescence emission of the beta-glucoronidase enzyme product 4-MU (4-methylumbelliferone) in a plate fluorescence lector, Spectra Max M3 apparatu (excitation 365 nm, emission 455 nm).
- After analysis defreezed samples can not be refreezed and are discarded.
- 4-Methylumbelliferone (4-MU) standards are used to calibrate the system.
- Protein concentration in the extracts is measured using the Bradford assay in a Spectra Max M3 apparatus.
- Embryos from each bombarded dish are frozen in liquid N2 and stored at -80 °C until analysis.
- Internal control in fluorometric studies, Two different internal controls are suggested
- Samples are cobombarded with a construction of luciferase enzyme fused to a constitutive promoter as maize ubiquitin (pUBI::LUC-nos-ter) to control homogenous bombardment of the different samples in an experiment.
- Freeze the embryos in liquid nitrogen after 24 h of bombardment, grind all the frozen tissue to a powder and resuspend by homogenization in 300 μl Luciferase lysis buffer (1x) previously tempered to room temperature.
- Sediment debris by brief centrifugation at 13,000 rpm with the MiniSpin Eppendorf Centrifuge at 4 °C and transfer supernatant to a new tube. Maintain on ice until analysis.
- Mix 10 μl of cell lysate with 100 μl of Luciferase Assay Reagent in a microtiter plate and quickly measure the light produced in a Spectra Max M3 apparatus.
- Make 40 μl aliquots with the remaining extract and freeze at -80 °C. Then proceed from step 6-e to do the fluorometric analysis of the studied promoters.
- Freeze the embryos in liquid nitrogen after 24 h of bombardment, grind all the frozen tissue to a powder and resuspend by homogenization in 300 μl Luciferase lysis buffer (1x) previously tempered to room temperature.
- Alternatively samples can be cobombarded with a mixture of two constructions that express two maize complementary transcriptional factors, the maize myb factor C1 (35S::I-C1) and the myc factor B-Peru (35S::I-B-Peru) involved in the anthocyanin biosynthesis. In these constructions the expression of these factors is under the control of the cauliflower mosaic virus (CAMV) 35S constitutive promoter and the first intron enhancer of the maize Adh1 (alcohol dehydrogenase 1). Expression of both factors results in red/bronze spots on the surface of the embryos that can be visually compared by observation with a stereomicroscope. Then proceed from step 6-a to do the fluorometric analysis of the studied promoters.
- Samples are cobombarded with a construction of luciferase enzyme fused to a constitutive promoter as maize ubiquitin (pUBI::LUC-nos-ter) to control homogenous bombardment of the different samples in an experiment.
Recipes
- Histochemical analysis detection buffer (10 ml)
1 ml sodium phosphate 1 M (pH 8.0)
27 mg (final concentration, 5 mM) Potassium hexacyanoferrate (III)
21 mg (final concentration, 5 mM) Potassium hexacyanoferrate (II)
6 μl Triton X-100
1.5 ml X-Gluc from stock (20 mg/ml in N,N-dimethylformamid) - Fluorometric lysis buffer
50 mM sodium phosphate (pH 7.0)
10 mM EDTA (pH 8.0)
10 mM BME (beta-Mercaptoethanol)
0.1% SDS (v/v)
0.1% Triton X-100 (v/v) - Fluorometric analysis reaction buffer (4 ml)
3 ml lysis buffer
1 ml methanol
1,764 mg MUG - Fluorimetric analysis stopping buffer
0.2 M Na2CO3 - MSO medium(1 L)
4.5 g M&S medium with vitamins (pH 5.8 with KOH 1 M)
30 g sucrose
2.4 g Gelrite - 1x Luciferase lysis buffer
25 mM Tris-phosphate (pH 7.8)
2 mM DTT
2 mM CDTA:1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid
10% glycerol
1% Triton® X-100
Acknowledgments
This protocol is adapted from Jose-Estanyol and Puigdomenech (2012).
References
- Jose-Estanyol, M. and Puigdomenech, P. (2012). Cellular localization of the embryo-specific hybrid PRP from Zea mays, and characterization of promoter regulatory elements of its gene. Plant Mol Biol 80(3): 325-335.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Jose-Estanyol, M. (2013). Maize Embryo Transient Transformation by Particle Bombardment. Bio-protocol 3(16): e865. DOI: 10.21769/BioProtoc.865.
Category
Plant Science > Plant transformation > Bombardment
Molecular Biology > DNA > Transformation
Cell Biology > Cell imaging > Fixed-tissue imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









