- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Metabolic Labeling of Yeast Sphingolipids with Radioactive D-erythro-[4,5-3H]dihydrosphingosine
Published: Vol 3, Iss 16, Aug 20, 2013 DOI: 10.21769/BioProtoc.862 Views: 10514
Reviewed by: Fanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Metabolite and Fatty Acid Analysis of Yeast Cells and Culture Supernatants
Liwei Chen and Wei Ning Chen
Sep 5, 2014 17859 Views

Sterol Analysis in Kluyveromyces lactis
Yvetta Gbelska [...] Alexandra Konecna
Sep 5, 2017 7801 Views

Integrated Membrane Yeast Two-Hybrid System for the Analysis of Membrane Protein Complexes
Brianna L. Greenwood [...] David T. Stuart
Aug 20, 2025 3036 Views
Abstract
Yeast sphingolipids can be metabolically labeled with exogenously added radioactive precursors. Here we describe a method to label ceramides and complex sphingolipids of Saccharomyces cerevisiae with a radioactive ceramide precursor, D-erythro-[4, 5-3H]dihydrosphingosine. This protocol is used to study the biosynthesis, transport and metabolism of sphingolipids in yeast.
Keywords: SphingolipidsMaterials and Reagents
- Yeast strain (Saccharomyces cerevisiae)
- D-erythro-[4,5-3H]-dihydrosphingosine ([3H]DHS) (60 Ci/mmol) (American Radiolabeled Chemicals, catalog number: ART-46 )
- Sodium fluoride (NaF) (Sigma-Aldrich, catalog number: S1504 )
- Sodium azide (NaN3) (Sigma-Aldrich, catalog number: S2002 )
- Glass beads (0.5 mm) (Yasui Kikai, catalog number: YGB05 )
- Chloroform (Sigma-Aldrich, catalog number: 05-3400 )
- Methanol (Sigma-Aldrich, catalog number: 19-2410 )
- 1-Butanol (Sigma-Aldrich, catalog number: 03-4560 )
- Ammonium hydroxide (Sigma-Aldrich, catalog number: 221228 )
- Acetic acid (Sigma-Aldrich, catalog number: 01-0280 )
- Thin layer chromatography silica gel 60 (TLC plate) (aluminium sheet 20 x 20 cm ) (Merck KGaA, catalog number: 105553 )
- N2 gas
- Distilled water (H2O)
- Dextrose (D-glucose) (Sigma-Aldrich, catalog number: 07-0680 )
- Yeast extract (Oriental Yeast, catalog number: OYC42008000 )
- Polypeptone (Wako Chemicals USA, catalog number: 513-76611 )
- Yeast nitrogen base without amino acids and ammonium sulfate (BD DifcoTM, catalog number: 233520 )
- Adenine (Sigma-Aldrich, catalog number: A9126 )
- Leucine (Nacalai Tesque, catalog number: 20327-62 )
- Histidine (Nacalai Tesque, catalog number: 18116-92 )
- Lysine (Sigma-Aldrich, catalog number: L5626 )
- Uracil (Nacalai Tesque, catalog number: 35824-82 )
- Ammonium sulfate (Sigma-Aldrich, catalog number: A2939 )
- Tryptophan (Nacalai tesque, catalog number: 35607-32 )
- Yeast extract-peptone dextrose (YPD) liquid medium (see Recipes)
- Synthetic minimal dextrose (SD) liquid medium (see Recipes)
- Water-saturated 1-butanol (see Recipes)
Equipment
- Tritium-sensitive imaging plate (Fujifilm, catalog number: BAS-TR2040 )
- Microcentrifuge tube
- Shaking incubator (TAITEC, model: BR-43FL.MR )
- Swinging bucket centrifuge (TOMY, model: LC-201 )
- Microcentrifuge (TOMY, model: MX-301 )
- Water bath shaker (TAITEC, model: PERSONAL-11 )
- Bath-type ultrasonic cleaner (AS ONE, model: US-2R )
- Pressure gas blowing concentrator (EYELA, model: MGS-2200 )
- TLC developing tank
- Hair dryer
- FLA-7000 imaging system (GE Healthcare Life Sciences, model: Typhoon FLA 7000 )
Software
- FLA-7000 image software (ImageQuant TL analysis)
Procedure
- Cell culture and metabolic labeling
- Inoculate yeast cells in YPD liquid medium at 25 °C with gyratory shaking at 175-200 rpm overnight.
- Dilute the culture with SD liquid medium to an OD600 of 0.01-0.02 and culture with gyratory shaking at 175-200 rpm overnight.
- When the OD600 of the culture is 0.2-0.6, transfer the culture to a 50 ml conical tube.
- Spin down yeast cells by a swinging bucket centrifuge at 2,000 x g for 5 min at room temperature (RT) and remove supernatant.
- Resuspend the pellet in 20 ml SD liquid medium, spin down at 2,000 x g for 5 min at RT and remove supernatant. Repeat this step at least three times.
- Resuspend cells in SD liquid medium to get an OD600 of 20 and transfer 0.5 ml of cell suspension to a new 50 ml conical tube.
- Incubate at 25 °C for 20 min with reciprocal shaking.
- Add 4 μCi of [3H] DHS to the cell culture and incubate at 25 °C for 1-4 h with reciprocal shaking. If necessary, lipid synthesis inhibitors are added before the labeling.
- Inoculate yeast cells in YPD liquid medium at 25 °C with gyratory shaking at 175-200 rpm overnight.
- Lipid extraction and alkaline hydrolysis
- To stop metabolic labeling, chill the 50 ml conical tube on ice and add 250 mM NaF and 250 mM NaN3 to a final concentration of 10 mM.
- Transfer the labeled culture to a 1.5 ml microcentrifuge tube (tube #1).
- Collect yeast cells by a microcentrifuge at 20,000 x g for 5 min at 4 °C, remove supernatant and resuspend yeast cells in 1 ml cold water. Repeat this step at least three times.
- Adjust volume of cell suspension to 66 μl with cold water and vortex well.
- Add 0.3 g of glass beads and vortex well at low temperature (vortex for 30 sec and chill on ice for 1-2 min, repeat at least three times).
- Add 440 μl of chloroform-methanol (CM; 1/1, v/v), vortex well and centrifuge at 20,000 x g for 5 min at RT.
- Transfer the supernatant to a new 1.5 ml microcentrifuge tube (tube #2).
- In order to extract the radiolabeled lipids remaining in tube #1, add 200 μl of chloroform-methanol-water (CMW; 10/10/3, v/v/v) in tube #1, sonicate for approximately 5-10 min in bath-type ultrasonic cleaner until the pellet is completely suspended, centrifuge at 20,000 x g for 5 min at RT and transfer the supernatant to tube #2.
- Dry the combined supernatants in tube #2 completely with N2 gas using pressure gas blowing concentrator.
- Add 80 μl of CMW, vortex well and centrifuge at 20,000 x g for 1 min at RT.
- To deacylate glycerophospholipids by mild alkaline hydrolysis, add 16 μl of 0.6 N NaOH in methanol, vortex well and incubate at 30 °C for 3 h.
- Centrifuge at 20,000 x g for 1 min at RT.
- To neutralize, add 16 μl of 0.6 N acetic acid in methanol, vortex well and centrifuge at 20,000 x g for 1 min at RT.
- Dry the reaction mixture completely with N2 gas using pressure gas blowing concentrator.
- To desalt, add 100 μl of water to tube #2, vortex well and spin down at 20,000 x g for 1 min at RT.
- Further add 200 μl of water-saturated 1-butanol, vortex well, centrifuge 20,000 x g for 5 min at RT and transfer the butanol (upper) phase containing sphingolipids to a new 1.5 ml microcentrifuge tube (tube #3).
- In order to collect the radiolabeled sphingolipids remaining in tube #2, add 200 μl of water-saturated 1-butanol to tube #2, vortex well, centrifuge 20,000 x g for 5 min at RT and transfer the butanol phase to tube #3. Repeat this step one more time.
- Add 100 μl of water to the combined butanol phase in tube #3, vortex well, centrifuge 20,000 x g for 5 min at room temperature and transfer the butanol phase to a new 1.5 ml microcentrifuge tube (tube #4).
- Dry the butanol phase in tube #4 completely with N2 gas using pressure gas blowing concentrator.
- To stop metabolic labeling, chill the 50 ml conical tube on ice and add 250 mM NaF and 250 mM NaN3 to a final concentration of 10 mM.
- Lipid separation and analysis
- Add 25 μl of CMW to tube #4, vortex well and centrifuge at 20,000 x g for 1 min at RT.
- Then load total sample on TLC plate.
- Place the plate in a glass TLC developing tank and develop with chloroform-methanol-4.2 N ammonium hydroxide (9/7/2, v/v/v) solvent mixture.
- When wetting front reaches within 1 cm of the top of TLC plate, remove the plate from the tank and dry it at RT.
- When the plate is completely dried with a hair dryer, set it in exposure cassette and expose it to a tritium-sensitive imaging plate for few hours-few days.
- Capture image and quantify signals with FLA-7000 image analyzer and software (Figure 1A).
- Add 25 μl of CMW to tube #4, vortex well and centrifuge at 20,000 x g for 1 min at RT.
- Ceramide extraction and analysis
- Add a few drops of water on the area containing ceramides of TLC plate, which is in 2 or 3 cm inside from the top edge of the plate.
- Collect the silica of the area by scraping with a spatula and transfer to a 1.5 ml microcentrifuge tube (tube #5).
- Add 400 μl of CM, sonicate for approximately 5 min in bath-type ultrasonic cleaner until the silica is completely suspended, centrifuge at 20,000 x g for 5 min at RT and transfer the supernatant to a new 1.5 ml microcentrifuge tube (tube #6).
- Add 200 μl of CM to tube #5, vortex well, centrifuge at 20,000 x g for 5 min at RT and transfer the supernatant to tube #6.
- Dry the combined supernatants in tube #6 completely with N2 gas using pressure gas blowing concentrator.
- Add 100 μl of water to tube #6, vortex well and spin down at 20,000 x g for 1 min at RT.
- Further add 200 μl of water-saturated 1-butanol, vortex well, centrifuge 20,000 x g for 5 min at RT and transfer the butanol phase containing ceramides to a new 1.5 ml microcentrifuge tube (tube #7).
- In order to collect the radiolabeled ceramides remaining in tube #6, add 200 μl of water-saturated 1-butanol to tube #6, vortex well, centrifuge 20,000 x g for 5 min at RT and transfer the butanol phase to tube #7. Repeat this step one more time.
- Dry the butanol phase in tube #7 completely with N2 gas usng pressure gas blowing concentrator.
- Add 25 μl of CMW to tube #7, vortex well and centrifuge at 20,000 x g for 1 min at RT.
- Then load total sample on TLC plate.
- Place the plate in a glass TLC developing tank and develop with chloroform-methanol-acetic acid (190/9/1, v/v/v) solvent mixture.
- When wetting front reaches within 1 cm of the top of TLC plate, remove the plate from the tank and dry it at RT.
- When the plate is completely dried with a hair dryer, set it in exposure cassette and expose it to a tritium-sensitive imaging plate for few hours-few days.
- Capture image and quantify signals with FLA-7000 image analyzer and software (Figure 1B).
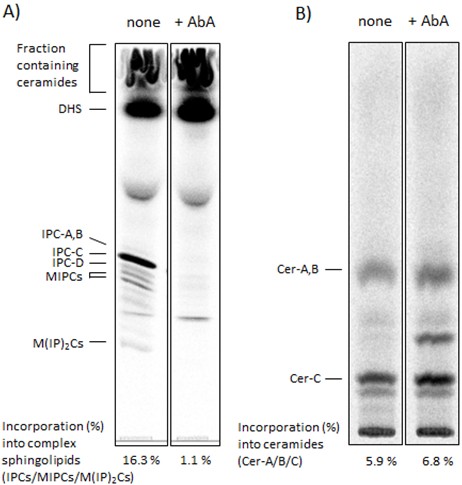
Figure 1. Captured images showing the distribution of complex sphingolipids (A) and ceramides (B) on TLC plates and quantified data. Wild-type cells were labeled with [3H]DHS at 25 °C for 3 h in the absence or presence of 2 μg/ml aureobasidin A (AbA), a specific inositol phosphorylceramide (IPC) synthase inhibitor (Funato and Riezman, 2001; Kajiwara et al., 2012). The labeled lipids were extracted, subjected to mild alkaline hydrolysis and analyzed by TLC with chloroform-methanol-4.2 N ammonium hydroxide (9/7/2, v/v/v) solvent mixture (A). Fractions containing ceramides in (A) (Funato and Riezman, 2001) were collected by scraping, and the radiolabeled ceramides were extracted from the silica and analyzed by TLC with chloroform-methanol-acetic acid (190/9/1, v/v/v) solvent mixture (B). Incorporation of [3H]DHS into complex sphingolipids (IPC-A,-B,-C,-D, MIPCs and M(IP)2Cs) and into ceramides (Cer-A, -B and -C) was quantified and determined as percentage of the total radioactivity in (A). IPC-A,-B,-C,-D and Cer-A,-B,-C are different IPC subclasses and ceramide species, respectively (Haak et al., 1997). The complex sphingolipids and ceramides can be identified by using mutants defective in the biosynthesis of specific sphingolipid species (Haak et al., 1997), by chemical treatment of isolated radiolabeled lipids (Puoti et al., 1991; Funato and Riezman, 2001) or by using the lipid standards that are isolated from [3H] inositol labeled sphingolipids (Puoti et al., 1991; Haak et al., 1997). MIPC, mannosyl inositolphosphorylceramide; M(IP)2C, mannosyl di(inositolphosphoryl)ceramide.
- Add a few drops of water on the area containing ceramides of TLC plate, which is in 2 or 3 cm inside from the top edge of the plate.
Recipes
- YPD liquid medium
20 g Dextrose
10 g Yeast extract
20 g Polypeptone
40 mg Uracil
40 mg Adenine
40 mg Tryptophan
Add H2O to 1 L
Sterilize by autoclaving.
- SD liquid medium
20 g Dextrose
1.7 g Yeast nitrogen base without amino acids and ammonium sulfate
5.0 g Ammonium sulfate
80 mg Uracil
80 mg Adenine
80 mg Leucine
80 mg Histidine
80 mg Lysine
Dissolve in 800 ml H2O
Adjust pH to 5.8-6.0 with 1 M NaOH
Adjust volume to 990 ml with H2O
Sterilize by autoclaving
Add 10 ml of tryptophan solution (8 mg/ml) sterilized by filtration.
- Water-saturated 1-butanol
Mix equal volumes of 1-butanol and H2O
Shake overnight at RT and leave
The resulting upper phase is water-saturated 1-butanol.
Acknowledgments
This protocol was adapted from Kajiwara et al. (2012). This work was supported by a grant from the Graduate School of Biosphere Science (Hiroshima University) to Kajiwara K. and by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science and from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to Funato K.
References
- Funato, K. and Riezman, H. (2001). Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J Cell Biol 155(6): 949-959.
- Haak, D., Gable, K., Beeler, T. and Dunn, T. (1997). Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J Biol Chem 272(47): 29704-29710.
- Kajiwara, K., Muneoka, T., Watanabe, Y., Karashima, T., Kitagaki, H. and Funato, K. (2012). Perturbation of sphingolipid metabolism induces endoplasmic reticulum stress-mediated mitochondrial apoptosis in budding yeast. Mol Microbiol 86(5): 1246-1261.
- Puoti, A., Desponds, C. and Conzelmann, A. (1991). Biosynthesis of mannosylinositolphosphoceramide in Saccharomyces cerevisiae is dependent on genes controlling the flow of secretory vesicles from the endoplasmic reticulum to the Golgi. J Cell Biol 113(3): 515-525.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Karashima, T., Kajiwara, K. and Funato, K. (2013). Metabolic Labeling of Yeast Sphingolipids with Radioactive D-erythro-[4,5-3H]dihydrosphingosine. Bio-protocol 3(16): e862. DOI: 10.21769/BioProtoc.862.
Category
Microbiology > Microbial metabolism > Lipid
Cell Biology > Cell metabolism > Lipid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









