- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fluorescence in situ Hybridization to the Polytene Chromosomes of Anopheles Mosquitoes
Published: Vol 3, Iss 16, Aug 20, 2013 DOI: 10.21769/BioProtoc.860 Views: 13545
Reviewed by: Fanglian HeYoko EguchiLin Fang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Preparation of Drosophila Polytene Chromosomes, Followed by Immunofluorescence Analysis of Chromatin Structure by Multi-fluorescence Correlations
Terra M. Kuhn [...] Maya Capelson
Jul 5, 2020 9932 Views

Labeling and Tracking Mitochondria with Photoactivation in Drosophila Embryos
Sayali Chowdhary and Richa Rikhy
Mar 5, 2022 4335 Views

Evaluation of Mitochondrial Turnover Using Fluorescence Microscopy in Drosophila
Felipe Martelli
Sep 5, 2022 2827 Views
Abstract
Fluorescence in situ hybridization (FISH) is a method that uses a fluorescently labeled DNA probe for mapping the position of a genetic element on chromosomes. A DNA probe is prepared by incorporating Cy-3 or Cy-5 labeled nucleotides into DNA by nick-translation or a random primed labeling method. This protocol was used to map genes (Sharakhova et al., 2010) and microsatellite markers (Kamali et al., 2011; Peery et al., 2011) on polytene chromosomes from ovarian nurse cells and salivary glands of malaria mosquitoes. Detailed physical genome mapping performed on polytene chromosomes has the potential to link DNA sequences to specific chromosomal structures such as heterochromatin (Sharakhova et al., 2010). This method also allows comparative cytogenetic studies (Sharakhova et al., 2011; Xia et al., 2010), and reconstruction of species phylogenies (Kamali et al., 2012).
Keywords: MosquitoMaterials and Reagents
- Early fourth instar Anopheles larvae
- Female Anopheles mosquitoes
- Template DNA
- Fisherfinest* Premium Extra-Thick Frosted Microscope Slides (Double frosted coating) (Thermo Fisher Scientific, catalog number: 12-544-6 )
- Fisherfinest* Premium Cover Glasses (22 x 22 mm) (Thermo Fisher Scientific, catalog number: 12-544-10 )
- 50% Propionic acid in water
- Razor blade
- Liquid nitrogen
- Ethanol, molecular biology grade
- Microscope slide staining jar with lid
- Random Primed DNA Labeling Kit (Roche Applied Science, catalog number: 11004760001 )
- Random Primers DNA Labeling System (Life Technologies, InvitrogenTM, catalog number: 18187-013 )
- Formamide (Super pure) (Fisher Bioreagent, catalog number: BP228-100 )
- Dextran Sulfate Sodium salt from Leuconostoc spp. (Sigma-Aldrich, catalog number: D8906 )
- Prolong® Gold antifade reagent (Life Technologies, InvitrogenTM, catalog number: P36930 )
- Cy3-dUTP (GE Healthcare, catalog number: PA53022 )
- Cy5-dUTP (GE Healthcare, catalog number: PA55022 )
- YOYO®-1 Iodide (491/509)–1 mM Solution in DMSO (Life Technologies, InvitrogenTM, catalog number: Y3601 )
- Paraformaldehyde (Sigma-Aldrich, catalog number: F8775 )
- DNA polymerase I (Fermentas, catalog number: EP0041 )
- DNase I (Fermentas, catalog number: EN0521 )
- QIAquick® Gel Extraction Kit (QIAGEN, catalog number: 28704 )
- QIAquick® PCR purification Kit (QIAGEN, catalog number: 28104 )
- Carnoy's solution (see Recipes)
- 20x SSC (see Recipes)
- 3 M NaAC (see Recipes)
- 1x PBS (see Recipes)
- Hybridization buffer (see Recipes)
Equipment
- 1.5 ml microcentrifuge tubes
- Forceps
- Disposable transfer pipette
- Dissecting needles
- Research stereo microscope (Leica, model: VA-OM-E194-354 )
- Phase contrast compound microscope with 10x, 20x, 40x and 100x objective lenses
- Thermal cycler
- Vacufuge® vacuum concentrator (Eppendorf, model: 022820001 )
- Incubator
- Water Bath
- Vortexer
- Confocal Microscope or Fluorescence Microscope
Procedure
- Polytene chromosome preparation
A-1 Salivary gland chromosome preparation- Preserve early fourth instar larvae in Carnoy's Solution and keep at -20 °C.
- Remove one fourth-instar larva from the vial with a pair of forceps and place it on a dust-free microscope slide with back upward, then put a drop of fresh Carnoy's solution onto it immediately (Figure 1a).
Note: Continue adding drops of Carnoy's solution when needed to prevent drying out until dipping 50% Propionic acid onto the gland.
While firmly holding the larva with one dissecting needle, gently pull the head away from thorax with another needle (Figure 1b). Insert a needle from the middle rear of the thorax just underneath the cuticle, and gently move forward to break the thorax cuticle along the mid dorsal line (Figure 1c). Carefully open up the thorax and separate the salivary gland from connecting tissue (Figure 1d and 1e). Remove the carcass and other tissue from slide and put one drop of fresh 50% Propionic acid onto the gland (Figure 1f).
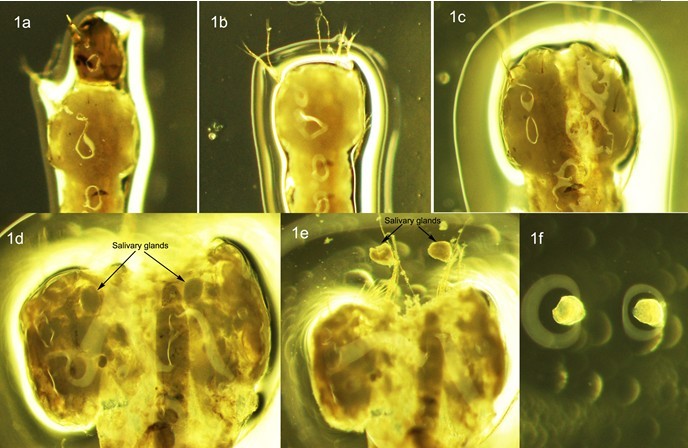
Figure 1. Dissection of salivary glands in 4th instar larva of An. Sinensis
- Cover gland with a dust-free coverslip and leave them for about 5 min. After 5 min, place a piece of filter paper over the coverslip, hold the four edges still with fingers. Gently tap it with a pencil eraser to release the polytene chromosomes from the salivary gland.
- Examine the banding pattern and spread of polytene chromosomes using a phase-contrast microscope.
- Place slides with good chromosomal preparations in a humid chamber with 4x SSC in the bottom of the chamber, at 60 °C for 15-20 min. After heating, put slides at 4 °C overnight or until immersing the slides in liquid nitrogen. Heating can be done on the Thermobrite machine with the absorbent strips soaked in distilled water.
Note: Slides can dry out if left at 4 °C for extended periods of time. Leaving them at 4 °C for longer than overnight is not recommended.
- While holding one corner of the slide with forceps or a gloved hand, dip chromosome preparation into the liquid nitrogen so that the coverslip is completely immersed. Hold slide in liquid nitrogen until the bubbling stops (usually 10-15 sec). Take it out of the liquid nitrogen and immediately remove the coverslip with a razor blade from one corner. It sometimes helps to put the slide on a flat surface when trying to remove the cover slip. Put slide in a slide jar with prechilled 50% Ethanol (-20 °C) and keep at 4 °C for at least 2 h.
- Dehydrate the preparations in slide jar with an ethanol series of 70%, and 90% for 5 min each at 4 °C and then 100% ethanol for 5 min at room temperature. Air dry, and keep slides in slide box until ready for use in in situ hybridization (Figure 2).
Note: Slides that are kept protected from dust and debris can be used for FISH at least within a year after the preparations are made.
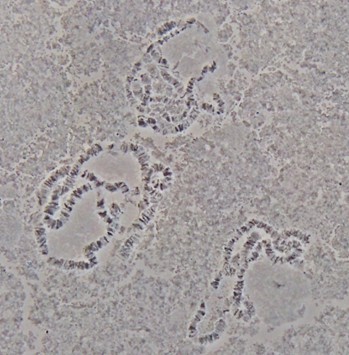
Figure 2. Polytene chromosomes from salivary glands of An. sinensis
- Dissect the ovaries of Anopheles mosquitoes from half gravid females 18-33 h after 2nd or 3rd blood feeding (Christophers' III stage) and keep 4-5 ovaries in a vial with 1 ml of Carnoy's sollution. After fixing the ovaries for 24 h at room temperate, transfer the vials to -20 °C for storage.
Note: Females should be bloodfed and lay eggs at least once before bloodfeeding again and dissecting ovaries for chromosomal preparations.
- To make the chromosome preparations, take one ovary out of the vials with a pair of forceps (or a transfer pipet) and place it into a drop of Carnoy's solution on microscope slide. After carefully removing tissues, trachea and blood, quickly separate the follicles from one ovary into 2-4 pieces. Up to four preparations can be made from one ovary.
Note: While dissecting, the ovaries should never be allowed to dry. Continue adding drops of Carnoy's solution when needed to prevent drying of the ovaries.
- On 4-8* microscope slides, add each of the pieces of divided ovary and one drop of 50% propionic acid on a separate slide. Let the pieces of ovary rest in propionic acid for about 5 min until follicles become clear, and swell to about twice their original size.
*number of slides you need depends of how many pieces each ovary is divided into.
- For each slide, use a dissecting microscope to separate the cleared follicles from each other and any other tissue or debris on the slide. Remove tissue and debris by wiping it away with a piece of paper towel, and apply a fresh drop of 50% propionic to the separated follicles.
- Do the same as steps 3-7 in "Salivary gland chromosome preparation".
- Preserve early fourth instar larvae in Carnoy's Solution and keep at -20 °C.
- Probe preparation and labeling
If using PCR products as probe, purify the PCR product from an agarose gel or from the PCR reaction using a QIAquick® Gel Extraction Kit or QIAquick® PCR purification Kit. Similar kits that remove excess nucleotides can also be used. However, when using a kit, dissolve the DNA in double distilled water instead of the elution buffer suggested in the final step.
B-1 Random Primer labeling protocol for fragments shorter than 1 kb (Random Primed DNA Labeling kit from Roche- Add 25 ng template DNA into double distilled water to a final volume of 13.5 μl in a microcentrifuge tube.
- Denature the DNA by heating in a boiling water bath for 10 min at 95 °C and chilling quickly in an ice bath.
- Add the following to the freshly denatured probes on ice:
Mix and centrifuge briefly.dGTP, 1.0 mM
1 μl
dCTP, 1.0 mM
1 μl
dATP, 1.0 mM
1 μl
Reaction Mixture (Vial 6)
2 μl
Klenow enzyme (Vial 7)
1 μl
Cy3 or Cy5-dUTP, 1.0 mM
0.5 μl
- Incubate for 1 h to 20 h (overnight) at 37 °C.
- Add 1/10 volume of 3 M NaAC and 2.5-3 volume of 100% ethanol. And mix by inverting the tubes. Keep at -80 °C or -20 °C for at least 3 h or until probes are needed for hybridization. If necessary, probes can be left in the freezer for long-term storage.
- Mix 1 μl DNA and 10 μl 2.5x Random Primer Solution and 2.5 μl sterile water well.
- Denature 5 min in boiling water or heating block, immediately cool on ice.
- Add 1.25 μl 1.0 mM dNTP mix (without a labeled dNTP), 8.75 μl water and 1 μl Klenow Fragment, mix gently but thoroughly.
- Add 0.5 μl Cy3 or Cy5-dUTP fluorescent nucleotide to each tube, when finished, tube must be covered immediately to protect from light. Mix well and incubate at 37 °C for 1.5 h.
- Do the same as step 5 in Section II-1 "Random Primer labeling protocol for fragments shorter than 1 kb (Random Primed DNA Labeling kit from Roche)".
- Prepare the following reaction mixture on ice:
Note: This protocol can be scaled down by 1/2 to accommodate 500 ng of template DNA.10x buffer for DNA Polymerase I
5 μl
1.0 mM dATP, dCTP, dGTP and 0.3 mM dTTP mixture
5 μl
DNase I freshly diluted to 0.02 units/μl
4 μl**
DNA Polymerase I
1 μl**
Template DNA
1 μg
Cy3- or Cy5-dUTP
1 μl
BSA diluted to 0.5 mg/ml
5 μl
Add water to final volume 50 μl
**Final concentrations of DNase I and DNA Polymerase I have to be optimized based on factors including initial size of template DNA, template DNA concentration and reaction time. Larger template size and greater template concentration generally require more DNase.
- Incubate the mix at 15 °C for 2-3 h.
- Run 3 μl of reaction mixture on an agarose gel to determine the size of digested fragments. Fragments should be 100-600 bp for best hybridization results. If fragments are still larger than this, incubate at 15 °C for additional time.
- To terminate the reaction and precipitate labeled probes, do the same as step 5 in Section B-2 "Random Primer labeling protocol for fragments shorter than 1 kb (Random Primed DNA Labeling kit from Roche)"
Note: Fluorescently labeled probes should be protected from light! In the steps following, even where it is not explicitly stated, make efforts to protect probes from light.
- Add 25 ng template DNA into double distilled water to a final volume of 13.5 μl in a microcentrifuge tube.
- Chromosomal fixation
- Do step C-2 and C-3 if slides are more than two months old. Otherwise, go to step C-4.
- Fix slides in 1:3 glacial acetic acid: methanol at RT for 10 min and air-dry.
- Dehydrate slides in 100% ethanol for 10 min and air dry again.
- Immerse slides in 1x PBS for 20 min at RT.
- Fix slides at room temperature in 4% paraformaldehyde for 1 min.
Note: Paraformaldehyde is hazardous and should be handled carefully. Avoid breathing gas or dust during preparation: wear gloves and other PPE when handling. Paraformaldehyde solution should not be dumped down drains.
- Dehydrate the slides through an ethanol series of 50%, 70%, 90%, 2x 100% for 5 min each at RT.
- Air-dry the slides.
- Do step C-2 and C-3 if slides are more than two months old. Otherwise, go to step C-4.
- In situ hybridization
- Centrifuge the tubes of labeled probes at 20,817 x g for 10 min. Carefully remove the supernatant and vacumfuge the tubes for 20 min to dry pellets.
- Dissolve dry probes in hybridization buffer prewarmed to 37 °C. The amount of hybridization buffer used to dissolve depends on the total amount of DNA you are dissolving. Dissolve 1 μg of DNA in 20-40 μl of warmed hybridization buffer.
- In a clean microcentrifuge tube, combine at least 250 ng each of one blue (Cy5 labeled) and one red (Cy3 labeled) probes. In situ hybridization is efficient if at least 500 ng of DNA is hybridized on the slide. Vortex and centrifuge the tube of combined probe briefly.
- Transfer the above prepared solution of combined probes to a chromosome preparation slide and cover with a 22 x 22 mm coverslip. Remove any large air bubbles with gentle pressure.
- Denature the target and probe DNA by placing the slides on the Thermobrite machine at 90 °C for 10 min. Thermobrite machine does not need to be humid.
- Seal edges of cover slip with rubber cement.
- Transfer the slides to pre-warmed humid chambers with 4x SSC at the bottom of the chambers and incubate at 39 °C for interspecies (e.g An. gambiae probe to An. stephensi chromosomes) or 42 °C for intraspecies hybridization for 3-18 h (usually overnight).
Note: Because there are fluorescently labeled probes on the slide, humid chambers should be impermeable to light.
- Centrifuge the tubes of labeled probes at 20,817 x g for 10 min. Carefully remove the supernatant and vacumfuge the tubes for 20 min to dry pellets.
- Washing
- Carefully remove rubber cement with forceps and coverslip.
- In a slide jar covered with aluminum foil, wash the slides with 1x SSC at 39 °C after interspecies or 0.2x SSC at 42 °C after intraspecies hybridization for 20 min in 50 ml without shaking.
- Wash the slides with 1x SSC after interspecies or 0.2x SSC after intraspecies hybridization at RT for 20 min in 50 ml without shaking.
- Dilute fluorescent dye YOYO-1 100 times in 1x PBS to make a stock solution. Mix 10 μl of 100x diluted YOYO-1 with 90 μl 1x PBS for each slide that you want to stain. The working solution of YOYO-1 is 1,000x diluted relative to original concentration.
- After washing in SSC for 20 min at room temperature, rinse slide in 1x PBS, and add 100 μl of YOYO-1 in PBS on each slide. Cover with parafilm. Leave at RT for 10 min inside of a slide box or somewhere dark.
- Rinse in 1x PBS and add 10 μl Prolong Gold antifade reagent, place coverslip on slide and blot out bubble. Keep in the slide box at 4 °C.
- Carefully remove rubber cement with forceps and coverslip.
- Signal detection
Detect the signals using a confocal or fluorescence microscope and map them to the polytene chromosomes of Anopheles mosquitoes (Figure 3).
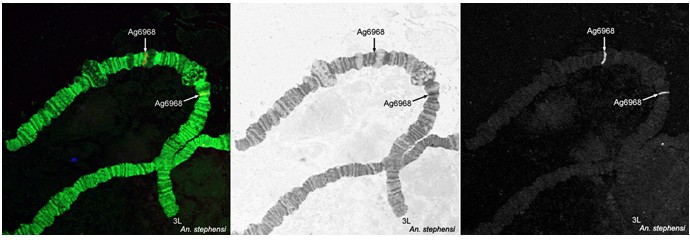
Figure 3. Fluorescence in situ hybridization and mapping of DNA probes on polytene chromosomes from ovarian nurse cells of An. stephensi
Recipes
- Carnoy's solution: Methanol:Glacial Acetic Acid = 3:1
Note: Carnoy's solution should be used with good ventilation or in a fume hood. Gloves should also be worn while using Carnoy's solution and it should not be disposed of down the drain.
- 1x PBS (1 L)
NaCl
8.01 g
KCl
0.20 g
NaH2PO4 (anhydrous)
1.15 g
KHP2O4 (anhydrous)
0.20 g
- 20x SSC (500 ml)
Sodium chloride
87.5 g
Sodium citrate
44 g
Add 1 N HCl to pH 7.0
- Hybridization buffer (2 ml)
20x SSC
120 μl
Dextran sulfate
0.2 g
Formamide
1.2 ml
Water
580 μl
- 3 M NaAC
Dissolve 24.61 g of Sodium Acetate (anhydrous) in 100 ml water.
Acknowledgments
The protocol was adapted from previously published papers: Sharakhova et al. (2010) and Kamali et al. (2012). We thank the Chinese Centre for Disease Control and Prevention, Shanghai, China for providing an Anopheles sinensis colony. This work was supported by a National Natural Science Foundation of China (31301877) to AX and by a National Institutes of Health (Bethesda, MD, U.S.A.) grant (5R21AI094289) to IVS. AP and IVS were supported in part by the Institute for Critical Technology and Applied Science (ICTAS) and the NSF award 0850198.
References
- Kamali, M., Sharakhova, M. V., Baricheva, E., Karagodin, D., Tu, Z. and Sharakhov, I. V. (2011). An integrated chromosome map of microsatellite markers and inversion breakpoints for an Asian malaria mosquito, Anopheles stephensi. J Hered 102(6): 719-726.
- Kamali, M., Xia, A., Tu, Z. and Sharakhov, I. V. (2012). A new chromosomal phylogeny supports the repeated origin of vectorial capacity in malaria mosquitoes of the Anopheles gambiae complex. PLoS Pathog 8(10): e1002960.
- Peery, A., M. V. Sharakhova, Antonio-Nkondjio, C., Ndo, C., Weill, M., Simard, F., and I. V. Sharakhov. 2011. Improving the population genetics toolbox for the study of the African malaria vector Anopheles nili: microsatellite mapping to chromosomes. Parasites and Vectors 4:202.
- Sharakhova, M. V., Xia, A., Tu, Z., Shouche, Y. S., Unger, M. F. and Sharakhov, I. V. (2010). A physical map for an Asian malaria mosquito, Anopheles stephensi. Am J Trop Med Hyg 83(5): 1023-1027.
- Sharakhova, M. V., George, P., Brusentsova, I. V., Leman, S. C., Bailey, J. A., Smith, C. D. and Sharakhov, I. V. (2010). Genome mapping and characterization of the Anopheles gambiae heterochromatin. BMC Genomics 11: 459.
- Sharakhova, M. V., Antonio-Nkondjio, C., Xia, A., Ndo, C., Awono-Ambene, P., Simard, F. and Sharakhov, I. V. (2011). Cytogenetic map for Anopheles nili: application for population genetics and comparative physical mapping. Infect Genet Evol 11(4): 746-754..
- Xia, A., Sharakhova, M. V., Leman, S. C., Tu, Z., Bailey, J. A., Smith, C. D. and Sharakhov, I. V. (2010). Genome landscape and evolutionary plasticity of chromosomes in malaria mosquitoes. PLoS One 5(5): e10592
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Xia, A., Peery, A., Kamali, M., Liang, J., Sharakhova, M. V. and Sharakhov, I. V. (2013). Fluorescence in situ Hybridization to the Polytene Chromosomes of Anopheles Mosquitoes. Bio-protocol 3(16): e860. DOI: 10.21769/BioProtoc.860.
- Kamali, M., Xia, A., Tu, Z. and Sharakhov, I. V. (2012). A new chromosomal phylogeny supports the repeated origin of vectorial capacity in malaria mosquitoes of the Anopheles gambiae complex. PLoS Pathog 8(10): e1002960.
Category
Cell Biology > Cell structure > Chromosome
Cell Biology > Cell imaging > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link