- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Pulmonary Myeloperoxidase Activity
Published: Vol 3, Iss 15, Aug 5, 2013 DOI: 10.21769/BioProtoc.851 Views: 10900
Reviewed by: Fanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Fluorescence Polarization-Based High-Throughput Screening Assay for Inhibitors Targeting Cathepsin L
Keyu Guo [...] Shuyi Si
Jul 20, 2025 2282 Views

Novel Experimental Approach to Investigate Immune Control of Vascular Function: Co-culture of Murine Aortas With T Lymphocytes or Macrophages
Taylor C. Kress [...] Eric J. Belin de Chantemèle
Sep 5, 2025 3551 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 496 Views
Abstract
Neutrophils are considered one of the first responders of the innate immune response. Their primary activities are to migrate to sites of infection by chemotaxis and trans-migration across the endothelium (Gaines et al., 2005). Once at the site of infection, they phagocytize microbes and kill them. Critical to the neutrophil’s ability to kill microbes are the multiple degradative enzymes contained within granules. The activity of these enzymes is non-specific, and therefore, neutrophils also contribute to tissue damage at the site of infection (Gaines and Berliner, 2005). Measurement of neutrophil infiltration into tissues is one way to gauge the severity of infection, inflammation, and tissue damage (Ayala et al., 2002). Myeloperoxidase is found in the primary granules of neutrophils and is an effective measure of neutrophil infiltration into tissues (Gaines and Berliner, 2005).
Keywords: NeutrophilMaterials and Reagents
- Fresh or snap-frozen tissues
- Liquid nitrogen
- MPO Fluoremetric Detection Kit (Assay Designs, catalog number: 907-029 )
- N-ethylmaleimide (Sigma-Aldrich, catalog number: E2068 )
- Hexadecyltrimethylammonium (Sigma-Aldrich, catalog number: 52366 )
- Deionized water
- DMSO (Sigma-Aldrich, catalog number: D8418 )
- Assay buffer (see Recipes)
- Hydrogen Peroxide (see Recipes)
- Detection Reagent (see Recipes)
- Reaction cocktail (see Recipes)
Equipment
- 14 ml Polystyrene test tubes
- 2 ml Microfuge tubes
- Black-welled 96 well plates
- Polytron homogenizer
- Sonicator
- Fluorescent plate reader
- Refrigerated centrifuge
Procedure
- Weigh out 50 mg tissue into polycarbonate test tubes containing 0.5-1 ml ice cold 1x assay buffer with 10 mM N-ethylmaleimide.
- Homogenize to just disrupt the tissue by placing the tip of the homogenizer in the bottom of the tube and switching the machine on, agitating the tube to move the tip of the homogenizer throughout the liquid and then switching the machine off after approximately one sec. Repeat until the tissues are just disrupted, usually 9 more times for a total of 10 pulses.
- Centrifuge at 500 x g for 10 min at 4 °C.
- Discard supernatant, add 500 μl ice cold 1x assay buffer containing 0.5% hexadecyltrimethylammonium to the pellet and transfer to a 2 ml microfuge tube.
- Homogenize to lyse the cells by placing the tip of the homogenizer in the bottom of the tube and switching the machine on, agitating the tube to move the tip of the homogenizer throughout the liquid and then switching the machine off after approximately 5 sec. Repeat 9 times for a total of 10 pulses. Place on ice.
- Sonicate to further lyse the cells at 50% power for 10 sec. Place on ice. Repeat 2 times for a total of 3 pulses.
- Snap freeze in liquid nitrogen and thaw at room temperature by placing in liquid nitrogen, immediately removing from the nitrogen and leave the samples at room temperature until completely thawed. Repeat snap freeze and thaw once.
- Store at -80 °C until assayed.
- Prior to assay, serially dilute the included MPO standard in assay buffer.
- To assay, add 50 μl of sample or the MPO standard to the bottom of a black 96 well plate. Add 50 μl Reaction Cocktail provided by the kit. All samples and standards should be run in duplicate.
- Incubate at room temperature in the dark for 30 min.
- Read the fluorescence at 550 nm excitation and 595 nm emission every 10 min until 60 min incubation.
- Plot the standard concentration vs. the maximum relative fluorescence units to create a standard curve (Figure 1).
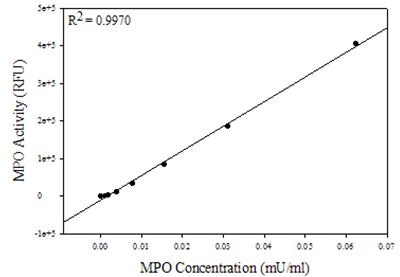
Figure 1. Representative Standard Curve
- Determine the concentration of the unknowns from the standard curve.
Recipes
- 1x Assay buffer
4 ml MPO assay buffer concentrate
36 ml deionized water
- Hydrogen Peroxide
22.7 μl 3% hydrogen peroxide
977 μl deionized water
- Detection Reagent
Supplied vial
500 μl DMSO
- Reaction cocktail
50 μl detection reagent
5 μl hydrogen peroxide
4.875 ml 1x assay buffer
Acknowledgments
This protocol is adapted from Ozment et al. (2012).
References
- Ayala, A., Chung, C. S., Lomas, J. L., Song, G. Y., Doughty, L. A., Gregory, S. H., Cioffi, W. G., LeBlanc, B. W., Reichner, J., Simms, H. H. and Grutkoski, P. S. (2002). Shock-induced neutrophil mediated priming for acute lung injury in mice: divergent effects of TLR-4 and TLR-4/FasL deficiency. Am J Pathol 161(6): 2283-2294.
- Gaines, P., Chi, J. and Berliner, N. (2005). Heterogeneity of functional responses in differentiated myeloid cell lines reveals EPRO cells as a valid model of murine neutrophil functional activation. J Leukoc Biol 77(5): 669-679.
- Ozment, T. R., Ha, T., Breuel, K. F., Ford, T. R., Ferguson, D. A., Kalbfleisch, J., Schweitzer, J. B., Kelley, J. L., Li, C. and Williams, D. L. (2012). Scavenger receptor class a plays a central role in mediating mortality and the development of the pro-inflammatory phenotype in polymicrobial sepsis. PLoS Pathog 8(10): e1002967.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Ozment, T. R. (2013). Pulmonary Myeloperoxidase Activity. Bio-protocol 3(15): e851. DOI: 10.21769/BioProtoc.851.
- Ozment, T. R., Ha, T., Breuel, K. F., Ford, T. R., Ferguson, D. A., Kalbfleisch, J., Schweitzer, J. B., Kelley, J. L., Li, C. and Williams, D. L. (2012). Scavenger receptor class a plays a central role in mediating mortality and the development of the pro-inflammatory phenotype in polymicrobial sepsis. PLoS Pathog 8(10): e1002967.
Category
Immunology > Immune cell function > Lymphocyte
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link