- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Western Blot Analysis of Chloroplast HSP70B in Chlorella Species
Published: Vol 3, Iss 15, Aug 5, 2013 DOI: 10.21769/BioProtoc.850 Views: 10939
Reviewed by: Ru Zhang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

MAPK Phosphorylation Assay with Leaf Disks of Arabidopsis
Pascale Flury [...] Sebastian Bartels
Oct 5, 2013 13897 Views

Use of a High-Affinity Ubiquitin-Binding Domain to Detect and Purify Ubiquitinated Substrates and Their Interacting Proteins
Nitu Saha [...] Mark Hochstrasser
Sep 5, 2025 3794 Views

Monitoring Endocytosis of Integral Membrane Proteins Using Western Blot-Based Detection of Biotinylated Antibody Uptake
Alexandra Graninger and Prasanna Satpute-Krishnan
Nov 20, 2025 2194 Views
Abstract
Western blotting allows for the specific detection of proteins by an antibody of interest. This protocol utilizes isolation of total proteins protocol for Chlorella vulgaris prior to gel electrophoresis. After electrophoresis, the selected antibodies are used to detect and quantify levels of chloroplast HSP70B.
Materials and Reagents
- Species
Three Chlorella species were used: C. vulgaris, isolated from soil samples of Livingston Island, the South Shetland Archipelago, Antarctic; C. vulgaris strain 8/1, isolated in 1968 from thermal springs in the region of Rupite, Bulgaria, and cultivated in our laboratory since 1975 and Chlorella kesslery a mesophile, from the Trebon collection.
Cultivation
Chlorella species were cultivated on TAP (Tris Acetate Phosphate) medium under continuous light of 60 μmol/m2/s and a temperature of 23 °C ± 0.1 °C in a Phytotron GC 400 growth chamber. The species were cultivated at this temperature because it is well known, that eurythermal algae, could be grown at a wide range of temperatures. - Rabbit polyclonal antibody HSP70B cytoplasmic (Agrisera, catalog number: AS06 175 )
- Goat anti-rabbit IgG(H&l) HRP conjugated (Agrisera, catalog number: AS09 602 )
- Coomassie brilliant blue G 250
- Orthophosphoric acid (Valerus, catalog number: N 4420 )
- Trichloroacetic acid (TCA)
- Bovine serum albumin (BSA) (Applichem GmbH, catalog number: 1391 0025 )
- Albumin fraction V (pH 7.0)
- Medium Pure Nitrocellulose (NCM) (0.45 μm) (Bio-Rad Laboratories, catalog number: 162-0115 )
- Filter paper
- Sponge
- 4CN (4-chloro-naphthol) (Bio-Rad Laboratories, catalog number: N170-6535 )
- N,N′ N′ Tetramethylethylendiamine (TEMED) (Alfa Aesar, catalog number: N12536 )
- Laemmli sample buffer (see Recipes)
- Reagent of Bradford (see Recipes)
- 5x Laemmli buffer (see Recipes)
- Running buffer (see Recipes)
- Transfer buffer (see Recipes)
- SDS-PAGE gel (see Recipes)
- 30% Acrylamide/N,N’-methylenebisacrylamide (AA/MBA) (see Recipes)
- 10% SDS (see Recipes)
- 10% Ammonium Persulfate (see Recipes)
- 1.5 M Tris HCl buffer (pH 8.8) (see Recipes)
- 1.0 M Tris HCl buffer (pH 6.8) (see Recipes)
- 4 M NaCl (see Recipes)
- 1.0 M Tris HCl buffer (pH 7.5) (see Recipes)
- 20% Tween 20 (see Recipes)
- Blocking buffer (see Recipes)
- Staining solution (see Recipes)
- 5% CH3COOH (see Recipes)
- Washing solution (see Recipes)
- 50 mM TBS-T buffer (see Recipes)
- HRP color development solution (see Recipes)
Equipment
- Motor
- Silica quartz sand 0.6 mm (Valerus, catalog number: N 1760 )
- Centrifuge (Sigma-Aldrich, model: 1-15 K)
- Electrophoresis chamber Transfer unit Hoefer miniVE electrophotesis and electrotransfer unit (Hoefer, model: SE300-10A-1.0 )
- Mini Rocker Shaker MR-1
Software
- Image J program
Procedure
- Cells lysis
- Add 100 μl Lisys Solution (LS) to the pellet (Chankova et al., 2013b) transfer to a chilled mortar, add two spatulas of silica sand, grind in the mortar for 3 min, add 200 μl LS in the mortar to wash and transfer the material into an Eppendorf tube of 2 ml.
- Centrifuge material from step 1 for 10 min at 14,500 x g.
- Separate the supernatant and heat the supernatant for 5 min at t = 90 °С.
- Centrifuge for 5 min at 14,500 x g.
- Split the supernatant in 2 samples: The first one use for the determination of protein concentration; the second one keep at t = -20 °С.
- Add 100 μl Lisys Solution (LS) to the pellet (Chankova et al., 2013b) transfer to a chilled mortar, add two spatulas of silica sand, grind in the mortar for 3 min, add 200 μl LS in the mortar to wash and transfer the material into an Eppendorf tube of 2 ml.
- Determination of protein concentration (Bradford)
- Add 30 μl 20% TCA to 30 μl supernatant.
- Centrifuge for 5 min at 14,500 x g.
- Add 60 μl 0.1 N NaOH to the pellet and mix thoroughly. To obtain best result add twice 30 μl of 0.1 N NaOH.
- Take 14 μl, add 86 μl 0.15 М NaCl and 3 ml reagent of Bradford.
- Use calibration curve for quantity of protein (Table 1).
For calibration curve:
Stock solution – 0.5 mg/ml BSA
Use Table 1 to determine every point of standard curve add 3 ml reagent of Bradford.
Table 1. Calibration curve for quantity of proteinN BSA (μg) BSA (0.5 mg/ml) vol (μl) NaCl (0.15 M) vol (μl) 1 0 0 100 2 5 10 90 3 10 20 80 4 15 30 70 5 20 40 60 6 25 50 50 7 30 60 40 8 35 70 30 9 40 80 20 10 45 90 10 11 50 100 0
- Add 30 μl 20% TCA to 30 μl supernatant.
- Protein electrophoresis
- Put about 100 ml of the 1x Laemmli buffer into cuvettes of electrophoresis module.
- Remove the comb and rinse the wells with buffer of SDS-PAGE gel.
- Pipet 10 μg protein into every well: adjust volumes so equal amount of protein is loaded (example: 10 μg protein are contained in 10 μl sample).
- Put the rest buffer in a bath of electrophoresis chamber (the volume must be always above minimum.).
- Run electrophoresis using the following parameters: 120 V and 16 mA for 3.5 h.
- When the electrophoresis is completed, remove the gel carefully.
Note: The order of the dropping of the samples. Concentrated gel should be released.
- Put about 100 ml of the 1x Laemmli buffer into cuvettes of electrophoresis module.
- Transfer of proteins on the NCM
- Soak the gel for 15 min in buffer.
- Soak sponge and filter paper for sandwich in transfer buffer.
- Cut NCM. The size should be such as the size of the gel. Put NCM for 5 min in transfer buffer.
Note: Mark the order of samples on the membrane! Label the membrane with a pencil. - Make a sandwich.
- The stack is assembled on the black cathode side (see Figure 1):
- Center a packing sponge on the black cathode side.
- Center a packing sponge on the black catode (a).
- Lay one piece of wet filter paper on the sponge (b).
- Position the equilibrate gels on the filter paper(c).
- Lay the membrane on the gel (d).
- Lay one piece of wet filter paper on the membrane (e).
- Lay two packing sponges on the filter paper (f).
- A second transfer stack if added, is placed between these two sponges.
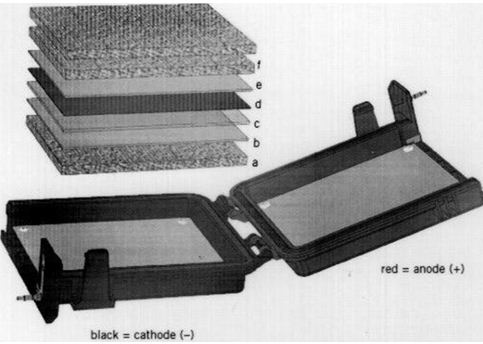
Figure 1. Assembling a transfer stack (this is an original figure taken from the Technical Guide available at www. hoeferinc.com)
- Center a packing sponge on the black cathode side.
- Different parts of the sandwich press very well, to avoid bubbles.
- Different parts of the sandwich should be well moistened. You can "roll" them with a tube.
- Close the apparatus. Put in a chamber transfer buffer.
- Run blotting with the following parameters: 35 V and 250 mA for 2 h.
- Close the apparatus. Put in a chamber transfer buffer.
- The stack is assembled on the black cathode side (see Figure 1):
- Soak the gel for 15 min in buffer.
- Western blot
- After the transfer of proteins, place the membrane in blocking buffer at t = 4 °C. Incubate on a rocker platform for 1 h (following this step we have obtained the best results).
- Place gels in staining solution for 4-5 h.
- Wash for 3-4 h the gel with washing solution.
- Dilute primary antibody in blocking buffer (1:10,000) and incubate according to manufacturer’s instructions. Incubate on a rocker platform at t = 4 °C overnight.
- Wash the membrane in TBS-T buffer on a rocker platform in a following way: twice for 2 min (2 х 2 min), after that twice for 10 min (2 x 10 min).
- Prepare secondary antibody in blocking buffer (1:20,000) and incubate according to manufacturer’s instructions. Incubate on a rocker platform at RT for 2 h.
- Wash the membrane in TBS-T buffer on a rocker platform in a following way: Twice for 2 min (2 х 2 min), after that three times for 5 min (3 x 5 min).
- Visualize using HRP Color Development Solution, 4CN according manufacturer’s instructions.
- Scan the membrane. Calculate protein amount using Image J program.
Recipes
- Laemmli sample buffer
2% SDS
5% 2-mercaptoehtanol
10% glycerol
0.002%(w/v) bromophenol blue
62.5 mM Tris HCl (pH 6.8) - Reagent of Bradford
Dissolve 100 mg Coomassie brilliant blue G 250 and 50 ml 96% alcohol in a stirrer for 15 min. Add 94.5 ml 90% orthophosphoric acid.
Add 900 ml deionized H2O and stir gently.
Filtering through a folded filter paper and make up to 1 L with deionized water.
Keep in a fridge at t = 4 °C. - 5x Laemmli buffer
15 g TRIS base
72 g Glycine in 1 L deionized H2O - Running Buffer
Add 200 ml 5x Laemmli buffer + 10 ml 10% SDS to 1 L deionized H2O - Transfer Buffer
Add 200 ml 5x Laemmli buffer + 2 ml 10% SDS to + 200 ml ethanol to 1 L deionized H2O - SDS-PAGE gel
Note: Glass tiles should be cleared well with alcohol before preparing SDS-PAGE gel.- Separating gel (12.5%) (Table 2)
Table 2. Preparing of separating gel solutionNumber of mini-gels 1 2 Deionized H2O 3.2 ml 6.4 ml Acrilamide/bisacrilamide (30%) 4 ml 8.0 ml 1.5 M Tris HCl buffer, pH 8.8 2.6 ml 5.2 ml 10% SDS 100 μl 200 μl 10% APS 100 μl 200 μl TEMED 10 μl 20 μl - Mix very carefully the components in a 50 ml Falcon tube to avoid bubbles.
- Insert separating gel between two glass plates of the chamber (about 1 cm below the boundary of tiles).
- Add deionized H2O carefully as a thin film using a syringe and wait about 15 min.
- Carefully remove the water; Wipe the water drops in the ends with filter paper.
- Mix very carefully the components in a 50 ml Falcon tube to avoid bubbles.
- Stacking gel (Table 3)
Table 3. Preparing of 4% stacking gel solutionNumber of mini-gels 1 2 Deionized H2O 1,370 μl 2,740 μl Acrilamide/bisacrilamide (30%) 330 μl 660 μl 1.0 M Tris HCl buffer, pH 6.8 250 μl 500 μl 10% SDS 20 μl 40 μl 10% APS 20 μl 40 μl TEMED 2 μl 4 μl - Put the concentrated gel, insert the comb and wait until the gel polymerize.
- For an electrophoresis is better to prepare about 1,250 ml 1x Laemmli buffer. It can be used twice.
- Separating gel (12.5%) (Table 2)
- 30% AA/MBA
29.0 g + 1.0 g MBA dissolve in 72.5 ml deionized H2O, make up the volume to 100 ml, filter using 0.45 μm filter
Keep at t = 4 °C less than 1 month. - 10% SDS
Dissolve 10 g SDS in 100 ml deionized H2O - 10% Ammonium Persulfate
Dissolve 1 g in 10 ml deionized H2O
Keep at t = 4 °C less than 1 month. - 1.5 M Tris HCl Buffer pH 8.8
Dissolve 18.5 g Tris base in 80 ml deionized H2O, adjust to pH = 8.8 with concentrated HCl and make up the volume to 100 ml. - 1.0 M Tris HCl Buffer pH 6.8
Dissolve 12.114 g Tris base in 80 ml deionized H2O, adjust to pH= 6.8 with concentrated HCl and make up the volume to 100 ml. - 50 mM TBS-T buffer
1.0 M Tris HCl buffer (pH 7.5)
200 mM NaCl
0.1% Tween 20 - 4 M NaCl
Dissolve 23.376 g NaCl in100 ml deionized H2O - 1.0 M Tris HCl buffer (pH 7.5)
Dissolve 12.114 g TRIS base in 80 ml deionized H2O, adjust to pH 7.5 with concentrated HCl and make up to the 100 ml. - 20% Tween 20
20 ml Tween make up to 100 ml deionized H2O. - Blocking buffer
Dissolve 5% fatless dry milk in 100 ml TBS-T buffer. - Staining solution
0.2% Coomassie Brilliant blue R- 250
40% C2H5OH - 5% CH3COOH
Dissolve 2 g Coomassie Brilliant blue R- 250, 400 ml C2H5OH and 50 ml CH3COOH and make up to 1 L with deionized H2O. - Washing solution
40% C2H5OH
5% CH3COOH - HRP Color Development Solution
Dissolve 60 mg of 4-chloro-naphtol into 20 ml of methanol.
Disolve immediately before use and protect solution from light.
Immediately prior to use, add 60 μl of ice cold 30% H2O2 to 100 ml TBS. Mix both solutions at RT. Use immediately.
References
- Chankova, S., Mitrovska, Z., Miteva, D., Oleskina, Y. P. and Yurina, N. P. (2013a). Heat shock protein HSP70B as a marker for genotype resistance to environmental stress in Chlorella species from contrasting habitats. Gene 516(1): 184-189.
- Chankova, S., Mitrovska, Z. and Yurina, N. (2013b). Heat Shock Treatment of Chlamydomonas reinhardtii and Chlorella Cells. Bio-protocol 3(15): e849.
- Chankova, S. G., Yurina, N. P., Dimova, E. G., Ermohina, O. V., Oleskina, Y. P., Dimitrova, M. T. and Bryant, P. E. (2009). Pretreatment with heat does not affect double-strand breaks DNA rejoining in Chlamydomonas reinhardtii. J Thermal Biol 34(7): 332-336.
- After the transfer of proteins, place the membrane in blocking buffer at t = 4 °C. Incubate on a rocker platform for 1 h (following this step we have obtained the best results).
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chankova, S., Mitrovska, Z. and Yurina, N. (2013). Western Blot Analysis of Chloroplast HSP70B in Chlorella Species . Bio-protocol 3(15): e850. DOI: 10.21769/BioProtoc.850.
Category
Plant Science > Phycology > Protein > Detection
Biochemistry > Protein > Immunodetection > Western blot
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












