- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Total RNA Isolation after Laser-capture Microdissection of Human Cervical Squamous Epithelial Cells from Fresh Frozen Tissue
Published: Vol 3, Iss 15, Aug 5, 2013 DOI: 10.21769/BioProtoc.848 Views: 11329
Reviewed by: Lin Fang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Mechanical Fractionation of Cultured Neuronal Cells into Cell Body and Neurite Fractions
Ankita Arora [...] J. Matthew Taliaferro
Jun 5, 2021 5499 Views

Updated Pseudo-seq Protocol for Transcriptome-Wide Detection of Pseudouridines
Yi Pan [...] Paul L. Boutz
May 5, 2024 1623 Views

An Optimized RNA Extraction Method From Micro-quantities of Guinea Pig Cartilage and Synovium for Osteoarthritis Research
Nidhi Bhardwaj [...] Jyotdeep Kaur
Jun 20, 2025 1618 Views
Abstract
As most tissue specimens contain a mixture of different cell types including epithelial cells, stromal cells and immune cells, selection of the cells of interest is of utmost importance for the accurate determination of gene/microRNA expression. Laser capture microdissection enables the researcher to obtain homogeneous ultrapure cell selections from heterogeneous starting material. The following protocol was optimized for the isolation of total RNA from cervical (premalignant) squamous epithelial cells from fresh frozen biopsy specimens.
Keywords: LCMMaterials and Reagents
- Fresh frozen tissue specimens stored in liquid nitrogen
- Tissue-Tek O.C.T. (Sakura Finetek Europe B.V., catalog number: 4583 )
- Mayers’ Haematoxylin
- Ethanol
- Xylene
- Liquid nitrogen or dry ice
- TRIzol (Life Technologies, catalog number: 15596-026 ) or a preferred alternative RNA isolation method
- Glycogen (F. Hoffmann-La Roche, catalog number: 10901393001 )
- Chloroform
- Isopropanol (Isopropyl alcohol)
- RNase-free water
Equipment
- 21-26 gauge needle
- PEN foil 2.0 UM covered slides (Leica, catalog number: 11505158 )
- 50 ml tubes with screw cap (Greiner Bio-one, catalog number: 210261 )
- Cryotome (rotary microtome in a frozen section environment)
- Leica Laser Microdissection system (AS LMD, model: LMD6500 or LMD7000 )
- Centrifuge
- -80 °C freezer for sample storage
Procedure
- Take all necessary precautions for RNA handling throughout the whole procedure.
- Wear gloves at all times.
- Only use RNase free equipment, plastics and reagents.
- Pretreat the PEN foil slides with UV light for 15 min to reduce static electricity.
Note: We use a UV source of 50 Hz and 26 Watt. - Cut 8-10 μm thick sections from the frozen tissue specimens (embedded in Tissue-Tek) and apply sections directly to the PEN foil covered side of the slide.
Notes:- More than one section can be applied to the same slide depending on the size of the specimen.
- Make sure that all sections are within the rectangle where the foil is not glued to the slide.
- Keep sections dry and cold at all times (store in a closed 50 ml tube at -80 °C and transport in liquid nitrogen or on dry ice).
- In addition a 4 μm section for standard H&E staining can be made to allow for demarcation of the tissue area/cells of interest by a pathologist or other expert.
- More than one section can be applied to the same slide depending on the size of the specimen.
- Slides containing tissue sections should be stored in a closed 50 ml tube and kept cold at all times (store at -80 °C and transport in liquid nitrogen or on dry ice).
Note: The use of 50 ml tubes avoids condensation on the slide thereby keeping the tissue dry which reduces RNA degradation. - Stain sections with Mayers’ Haematoxylin for 1 min at room temperature (RT).
Notes:- Before staining let slides come to RT while still in the closed 50 ml tube.
- Be careful not to lose the tissue section. Staining is best done by pipetting the haematoxylin directly on the slide.
- Before staining let slides come to RT while still in the closed 50 ml tube.
- Carefully rinse slide in sterile (RNase free) water.
- Dehydrate tissue sections by subsequent rinsing for 1 min each in 50%, 70% and 100% ethanol (can be done in 50 ml tubes) and an 8 min incubation in Xylene (use a glass container).
- Store slides in a closed 50 ml tube at -80 °C or continue with the microdissection.
- Add a little (10-20 μl) TRIzol to the tube cap in which the microdissected tissue will fall.
Notes:- Let slides come to room temperature while in the 50 ml tube right before starting the microdissection procedure for that slide.
- If an alternative RNA isolation method is preferred, TRIzol should be substituted by the first reagent used in that method.
- Let slides come to room temperature while in the 50 ml tube right before starting the microdissection procedure for that slide.
- Microdissect all areas/cells of interest from the slide using at a 10x magnification (see Figure 1) using the following settings:
Intensity: 46
Speed: 5 (or lower)
Offset: 5
Aperture diff: 6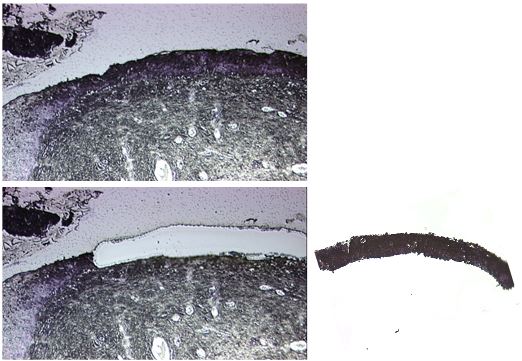
Figure 1. Example picture of a cervical tissue section before (upper panel) and after (lower panel) laser capture microdissection of the dysplastic epithelium. The microdissected tissue in the cap is shown in the left panel. - Add another 80-90 μl of TRIzol (or alternative isolation reagent) to the 0.2 ml tube containing the microdissected tissue and transfer tissue and TRIzol/alternative isolation reagent to a 1.5 ml tube.
Note: If preferred by the researcher an alternative RNA isolation method can be used. However, this protocol was optimized using TRIzol. - Add TRIzol to the tube to obtain a total volume of 1 ml.
Note: Store at -80 °C or continue with RNA isolation. - Add 1 μl glycogen and mix well (vortex for 10 seconds).
- Disrupt the tissue/cells and shear the genomic DNA with 10 passes through a 21-26 gauge needle.
- Add 0.2 ml of chloroform and vortex for 30 sec.
- Spin down for 5 min at full speed (12,000 x g) at RT.
- Transfer the aqueous phase to a clean tube.
- Add 0.5 ml of isopropanol to the aqueous phase and mix well.
- Incubate samples for at least 1 h at -20 °C for optimal precipitation of the RNA.
- Spin down for 15 min at full speed (12,000 x g) at RT.
- Discard supernatant
Note: Careful not to disturb the pellet containing your RNA. - Wash pellet with 200 μl of 70% ethanol.
- Spin down at full speed for 10 min at RT.
- Discard supernatant.
- Repeat steps 20-22.
Note: This time it is important to completely remove the supernatant. - Airdry the pellet.
Note: If all supernatant in removed in step 23 this should only take a few minutes. - Resuspend the pellet in 10-30 μl RNase free water.
- Store at -80 °C.
Acknowledgments
This protocol was optimised with the help of Elza de Bruin (de Bruin et al., 2005) and Muriel Verkuijten. This work was supported by the Centre for Medical Systems Biology (CMSB) in the framework of the Netherlands Genomic Initiative, Royal Netherlands Academy of Arts and Sciences, the VUMC-CCA institute of the VU University Medical Center, Amsterdam, The Netherlands (grant number CCA20085-04) and the Dutch Cancer Society (KWF, grant number VU2010-4668).
References
- de Bruin, E. C., van de Pas, S., Lips, E. H., van Eijk, R., van der Zee, M. M., Lombaerts, M., van Wezel, T., Marijnen, C. A., van Krieken, J. H., Medema, J. P., van de Velde, C. J., Eilers, P. H. and Peltenburg, L. T. (2005). Macrodissection versus microdissection of rectal carcinoma: minor influence of stroma cells to tumor cell gene expression profiles. BMC Genomics 6: 142.
- Wilting, S. M., de Wilde, J., Meijer, C. J., Berkhof, J., Yi, Y., van Wieringen, W. N., Braakhuis, B. J., Meijer, G. A., Ylstra, B., Snijders, P. J. and Steenbergen, R. D. (2008). Integrated genomic and transcriptional profiling identifies chromosomal loci with altered gene expression in cervical cancer. Genes Chromosomes Cancer 47(10): 890-905.
- Wilting, S. M., Snijders, P. J., Verlaat, W., Jaspers, A., van de Wiel, M. A., van Wieringen, W. N., Meijer, G. A., Kenter, G. G., Yi, Y., le Sage, C., Agami, R., Meijer, C. J. and Steenbergen, R. D. (2013). Altered microRNA expression associated with chromosomal changes contributes to cervical carcinogenesis. Oncogene 32(1): 106-116.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wilting, S. M. and Steenbergen, R. D. (2013). Total RNA Isolation after Laser-capture Microdissection of Human Cervical Squamous Epithelial Cells from Fresh Frozen Tissue . Bio-protocol 3(15): e848. DOI: 10.21769/BioProtoc.848.
Category
Molecular Biology > RNA > RNA extraction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









