- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of Endogenous MALT1 Activity
Published: Vol 3, Iss 14, Jul 20, 2013 DOI: 10.21769/BioProtoc.821 Views: 12826
Reviewed by: Lin FangFanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measuring Protein Synthesis during Cell Cycle by Azidohomoalanine (AHA) Labeling and Flow Cytometric Analysis
Koshi Imami and Tomoharu Yasuda
Apr 20, 2019 9752 Views

Analysis of the Ubiquitination and Phosphorylation of Vangl Proteins
Di Feng [...] Bo Gao
Oct 20, 2022 3422 Views

Isoform-specific, Semi-quantitative Determination of Highly Homologous Protein Levels via CRISPR-Cas9-mediated HiBiT Tagging
Kristina Seiler [...] Mario P. Tschan
Jul 20, 2023 2521 Views
Abstract
MALT1(Mucosa associated lymphoid tissue protein 1) is an important adapter protein for the NF-kB driven lymphocyte activation and the development and survival of distinct B-cell lymphoma entities. In addition MALT1 is a cysteine protease that structurally resembles caspases while having a different substrate preference and mechanism of activation. This paracaspase activity of MALT1 has been shown to be critical for an optimal NF-kB activation and survival of the aggressive ABC-DLBCL (Activated B cell-type of diffuse large B cell lymphoma), which highlights the protease as an attractive therapeutic target for the treatment of distinct B-cell lymphomas and immune diseases like rheumatoid arthritis or multiple sclerosis. In this protocol we describe a fluorogenic cleavage assay, which can be used to measure endogenous and also ectopic MALT1 activity. To this end, cellular MALT1 needs to be precipitated from the lysed cells via antibody immunoprecipitation and subsequently incubated with a fluorogenic substrate peptide. The MALT1 cleavage assay has been developed to directly determine the activity profile of MALT1 in the course of the adaptive immune response as well as in pathological signaling in lymphoid malignancies. In addition, the MALT1 activity assay has been successfully used to monitor cellular MALT1 inhibition with small molecule inhibitors.
Keywords: ProteaseMaterials and Reagents
- Primary human and murine cells
- T-cell-lines (Jurkat)
- B-cell-lines: ABC-(TMD8, HBL1, OCI-Ly3, U2932, OCI-Ly10, RIVA) and GCB-DLBCL (Su-DHL-6, Su-DHL-4, BJAB)
- RPMI 1640 (Life Technologies, catalog number: 21875 )
- IMDM (Life Technologies, catalog number: 21056 )
- Fetal Bovine Serum (FBS) (Life Technologies, catalog number: 10270 )
- PMA (Phorbol 12-Myristat 13-Acetat) (Calbiochem, catalog number: 16561-29-8 )
- Lonomycin (Calbiochem, catalog number: 407950 )
- Anti-CD3/CD28 (Hit3a/CD28.2) (BD Heidelberg)
- IgG1 (R19-15) (BD Pharmingen, catalog number: 553440 )
- IgG2a (A85-1) (BD Pharmingen, catalog number: 553387 )
- MALT1 antibody (Santa Cruz, catalog number: H300 )
- Protein-G Sepharose (GE Healthcare, catalog number: 17-0618-01 )
- PBS (Life Technologies, catalog number: 14190 )
- Ac-LRSR-AMC (Peptides International, catalog number: MCA-3952-PI )
- Z-VRPR-FMK (ENZO Life Sciences, catalog number: ALX-260-166 )
- Complete protease inhibitor cocktail tablets (Roche, catalog number: 11836145001 )
- Mepazine (Hit2lead, catalog number: 5216177 )
- 384-well non-binding plates, black (Greiner Bio-one, catalog number: 781900 )
- Saccharose
- CHAPS
- HEPES
- Triton X-100
- Cellular lysis buffer (see Recipes)
- MALT1 cleavage buffer (see Recipes)
Equipment
- 37 °C 5% CO2 cell culture incubator
- 26 G syringe (Roth, catalog number: C718.1 )
- Centrifuge (Eppendorf, model: 5417R )
- Synergy II multiwell plate reader (Biotek)
- Rotary Mixer
Procedure
- To activate MALT1 plate 2.5 x 106 cells (Jurkat T-cells, primary T cells or PBMCs) and either stimulate with PMA/Ionomycin (200 ng/ml and 300 ng/ml final concentrations) in 10 ml of complete RPMI media in 25 cm2 cell culture flasks or with anti-CD3 and anti-CD28 antibodies (1 μg/ml final concentration each) in the presence of anti-IgG1/IgG2a coupling antibodies (0.5 μg/ml each) in 1 ml of complete RPMI media in 12-well for 30 min or treat with solvent (H2O) in the control. Cells with constitutive MALT1 activity (e.g. ABC-DLBCL) can be left untreated.
- As a negative control treat the above cells with MALT1 inhibitors, e.g. the tetrapeptide Z-VRPR-FMK (50 μM) or small molecule inhibitors like mepazine (10 μM) 3 - 6 h prior to stimulation and lysis.
- All 2.5 x 106 cells per reaction are then pelleted at 300 x g for 5 min.
- Removal of supernatant and lysis of the cells with cellular lysis buffer (500 μl) by rotating for 20 min on 4 °C in a 1.5 ml reagent tube.
- Centrifugation of the lysates for 10 min at 21,000 x g to remove cell debris.
- Incubation of all of the lysate with 700 ng MALT1 antibody over night at 4 °C on a rotary mixer.
- Incubation with 15 μl Protein-G sepharose beads (beads were washed and diluted 1:2 in PBS and equilibrated before usage) for 60 min at 4 °C on a rotary mixer.
- Beads are pelleted and washed 3 times with PBS at 300 x g for 2 min at 4 °C.
- In the last washing step all PBS is discarded via a syringe and 45 μl cleavage buffer is added to the beads.
- The beads are pelleted at 300 x g for 1 min afterwards re-suspended carefully and 49 μl of the suspension (beads and cleavage buffer) are subsequently transferred to one well of a 384-well assay plate.
- 1 μl of the fluorogenic substrate Ac-LRSR-AMC is then added to the wells in a final concentration of 20 μM and the plates are placed into the plate reader where they are shaked for 10 sec to mix the reagents.
- After an incubation of 30 min at 30 °C the release of AMC fluorescence due to MALT1 cleavage is measured at 360 nm excitation and 460 nm emission over a time course of 90 min.
- Specificity of MALT1 cleavage activity in vitro can be assessed by adding 5 nM of Z-VRPR-FMK to a separate control reaction (negative control).
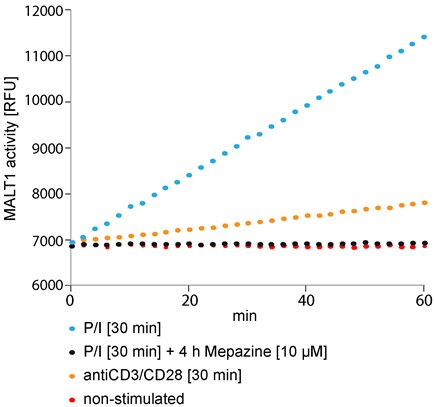
Figure 1. Fluorogenic MALT1 cleavage assay. Jurkat T-cells (2.5 x 106) were pre-incubated with Mepazine or DMSO for 4 h and subsequently stimulated with PMA/Ionomycin (P/I) or antiCD3/CD28 for 30 min or left untreated. After lysis of the cells MALT1 was precipitated and the catalytic activity was measured after addition of Ac-LRSR-AMC in a Synergy II plate reader over 60 min at 2 min intervals.
Recipes
- Cellular lysis buffer
50 mM HEPES (pH 7.5)
10% (v/v) Glycerin
0.1% (v/v)Triton X-100
1 mM Dithiothreitol (DTT)
150 mM NaCl
2 mM MgCl2
1 complete protease inhibitor tablet per 50 ml
- MALT1 cleavage buffer
50 mM MES (pH 7.0)
150 mM NaCl
10% (w/v) Saccharose
0.1% (w/v) CHAPS
1 M Sodium citrate
10 mM DTT
Acknowledgments
This protocol was adapted from two previous publications (Kloo et al., 2011; Nagel et al., 2012). The work was supported by a grant of the ‘Deutsche Krebshilfe e.V.’ to DK.
References
- Kloo, B., Nagel, D., Pfeifer, M., Grau, M., Duwel, M., Vincendeau, M., Dorken, B., Lenz, P., Lenz, G. and Krappmann, D. (2011). Critical role of PI3K signaling for NF-kappaB-dependent survival in a subset of activated B-cell-like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci U S A 108(1): 272-277.
- Nagel, D., Spranger, S., Vincendeau, M., Grau, M., Raffegerst, S., Kloo, B., Hlahla, D., Neuenschwander, M., Peter von Kries, J., Hadian, K., Dorken, B., Lenz, P., Lenz, G., Schendel, D. J. and Krappmann, D. (2012). Pharmacologic inhibition of MALT1 protease by phenothiazines as a therapeutic approach for the treatment of aggressive ABC-DLBCL. Cancer Cell 22(6): 825-837.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Nagel, D. and Krappmann, D. (2013). Measurement of Endogenous MALT1 Activity. Bio-protocol 3(14): e821. DOI: 10.21769/BioProtoc.821.
Category
Cancer Biology > General technique > Biochemical assays > Protein analysis
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










