- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Motility Assay for Zebrafish Embryos
Published: Vol 3, Iss 11, Jun 5, 2013 DOI: 10.21769/BioProtoc.787 Views: 12574
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Reproducible Protocol to Measure the Critical Swimming Speed of Adult Zebrafish
Yuma Wakamatsu [...] Hiromi Hirata
Aug 20, 2020 5680 Views

Automated Analysis of Cerebrospinal Fluid Flow and Motile Cilia Properties in The Central Canal of Zebrafish Embryos
Olivier Thouvenin [...] Claire Wyart
Mar 5, 2021 6481 Views

Live Imaging Transverse Sections of Zebrafish Embryo Explants
Eric Paulissen and Benjamin L. Martin
Feb 5, 2024 1896 Views
Abstract
Analyzing the swimming ability of 2 days post fertilization zebrafish embryos can be a useful technique to study neuromuscular function. Here is a protocol for determining the time it takes for zebrafish embryos to swim a predetermined distance.
Keywords: MuscleMaterials and Reagents
- Pipette pump
- Glass Pasteur pipette
- Overhead transparency sheet
- Zebrafish embryo media of choice (Goody et al., 2012)
- Embryo ‘poker’ tool (e.g. segment of fishing line super glued in the end of a glass capillary tube, fire polished glass rod)

Figure 1. Embryo poker tool. This is a useful tool for the positioning and touch stimulus of zebrafish embryos. Fishing line (10 lb, 0.012 inch diameter) is super glued into the end of a glass capillary tube (Sutter Instruments, catalog number: BF100-50-10) with approximately 1 cm of overhang. The glass capillary tube is then wrapped in lab tape.
Equipment
- Microscope with high-speed, digital video camera attachment (e.g. Zeiss SteREO Discovery.V12 with Zeiss Axiocam HSm)
- Video processing software (e.g. Zeiss Axiovision)
- 60 mm Petri dish
Procedure
A. Preparation:
- Print the motility wheel (Figure 2) on an overhead transparency sheet.
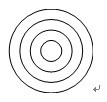
Figure 2. Motility wheel. The diameters of these concentric circles provide predetermined distances for zebrafish embryos to swim. The diameters of the circles are, in ascending order: 5 mm, 10 mm, 15 mm, 20 mm.
- Place the overhead transparency sheet on the stage of the microscope that will be used to record the videos and adjust the magnification of the microscope such that the edges of the desired concentric circle (e.g. the circle with a 10 mm diameter) are just within the field of view. Do not readjust the magnification setting within a motility assay experiment and use this magnification setting for any subsequent replicates that will be pooled together or compared.
- Center a 60 mm Petri dish containing zebrafish embryo media over the concentric circles on the overhead transparency sheet on the microscope stage.
- Using a pipette pump and glass Pasteur pipette, transfer one embryo into the Petri dish and use a poker to gently move the embryo into the middle of the concentric circles.

Figure 3. Microscope set up. A 60 mm Petri dish containing embryo media is placed on top of the motility wheel on top of the microscope stage. An embryo poker tool is used to position a zebrafish embryo in the middle of the motility wheel. Photo credit: University of Maine, Mike Mardosa.
B. Video acquisition:
- Begin recording a video when the embryo is stationary and in the center of the concentric circles.
- Looking through the eyepieces of the microscope, gently poke the tail of the embryo with a poker. Ensure that you are holding the poker such that your hand does not appear in the video.
- When the embryo completely exits the predetermined concentric circle, stop the video recording. A normal 2 day old embryo will exit the circle with a 10 mm diameter in approximately 200 milliseconds on the first poke (Goody et al., 2012).
- If the embryo does not completely exit the designated circle, use the poker to reposition the embryo in the center of the circles and repeat steps B1-3. After multiple unsuccessful attempts (~10 attempts), it may be determined that the embryo is incapable of exiting the circle and video recording can be stopped.
- Repeat these video acquisition steps until videos of all the desired embryos have been recorded.
C. Video analysis:
- ‘Cut’ the videos to only contain the necessary frames, if desired. Save the videos.
- For each video, scroll through the frames and determine the first frame in which the entire body of the embryo has exited the predetermined circle. Record that frame number in a spreadsheet. Scroll back to the beginning frames of that same video and determine the last frame in which the embryo is stationary prior to the touch stimulus. Record that frame number in the spreadsheet.
- Repeat step C2 for all the videos.
- Determine the time lag that occurs between each of the frames (e.g. 9 ms).
- Calculate the time it takes an embryo to swim a predetermined distance in milliseconds by subtracting the beginning frame number from the end frame number and multiplying by the time lag value. Record this value in the spreadsheet.
- Repeat step C5 for all the motility videos.
- After all the biological replicates for a motility experiment have been completed and analyzed, assign the greatest (i.e. slowest) value recorded within a treatment group to all the embryos in that same treatment group that never successfully exited the designated circle.
- Calculate the mean and standard deviation or standard error of the mean for each treatment group and statistically compare the values using an unpaired student’s t-test with unequal variance. For an example experimental result using this motility assay, please refer to (Goody et al., 2012).
Acknowledgments
This protocol was adapted from the previous publication: Goody et al. (2012). Development of this protocol was supported by NIH grant RO1 HD052934-01A1 to CAH. MFG would like to thank the University of Maine Graduate School of Biomedical Sciences and Engineering for funding.
References
- Goody, M. F., Kelly, M. W., Reynolds, C. J., Khalil, A., Crawford, B. D. and Henry, C. A. (2012). NAD+ biosynthesis ameliorates a zebrafish model of muscular dystrophy. PLoS Biol 10(10): e1001409.
- Westerfield, M. (1993) The Zebrafish Book: A Guide for the Laboratory Use of the Zebrafish (Brachydanio rerio) University of Oregon Press, Eugene.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Goody, M. F. and Henry, C. A. (2013). Motility Assay for Zebrafish Embryos. Bio-protocol 3(11): e787. DOI: 10.21769/BioProtoc.787.
Category
Developmental Biology > Morphogenesis > Motility
Neuroscience > Sensory and motor systems
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link