- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection of Membrane Potential in Mycobacterium tuberculosis
Published: Vol 3, Iss 11, Jun 5, 2013 DOI: 10.21769/BioProtoc.785 Views: 15561

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measurement of Extracellular Ca2+ Influx and Intracellular H+ Efflux in Response to Glycerol and PEG6000 Treatments
Tao Li and Baodong Chen
Sep 20, 2013 10234 Views

Bacterial Intracellular Sodium Ion Measurement using CoroNa Green
Yusuke V. Morimoto [...] Tohru Minamino
Jan 5, 2017 10445 Views

Live Cell Measurement of the Intracellular pH of Yeast by Flow Cytometry Using a Genetically-Encoded Fluorescent Reporter
Catherine G. Triandafillou and D. Allan Drummond
Jun 20, 2020 4826 Views
Abstract
DiOC2 (Novo et al., 2000) exhibits green fluorescence in all bacterial cells, but the fluorescence shifts towards red emission as the dye molecules self associate at the higher cytosolic concentrations caused by larger membrane potentials. Proton ionophores such as CCCP destroy membrane potential by eliminating the proton gradient. The magnitude of membrane potentials varies with different bacterial species. For many gram-positive species, including Staphylococcus aureus and Micrococcus luteus, the red:green ratio tends to vary with the intensity of the proton gradient while in many gram-negative bacteria such as Escherichia coli and Salmonella choleraesuis, the response of the dye does not appear to be proportional to proton gradient intensity. Mycobacterium tuberculosis itself is a difficult organism to work with because of its rigid cell wall.
Materials and Reagents
- Mycobacterial cells (Mycobacterium tuberculosis)
- 7H9 broth (Difco, catalog number: 271310 )
- Glycerol (Amresco, catalog number: 0854 )
- TWEEN®80 (Amresco, catalog number: 0442 )
- Middlebrook ADC (albumin-dextrose-catalase) enrichment (BD Biosciences, catalog number: 211886 )
- The BacLightTM Bacterial Membrane Potential Kit (Invitrogen, catalog number: B34950 )
Provides the following solution:- DiOC2 (Component A), 1.2 ml of a 3 mM solution in DMSO
- CCCP (Component B), 300 μl of a 500 μM solution in DMSO
- Phosphate-buffered saline (PBS, Component C), 10 mM sodium phosphate and 145 mM sodium chloride, pH 7.4
- DiOC2 (Component A), 1.2 ml of a 3 mM solution in DMSO
- Supplenmented 7H9 broth (see Recipes)
Equipment
- Incubation shaker
- Laminar flow hood
- Laboratory centrifuge
- Flow cytometer (BD FACSVerseTM System)
Procedure
- Culture the mycobacterial cells aerobically in 7H9 broth at 37 °C with shaking (200 rpm) till mid- log phase (OD600 of 0.3).
- Filter the required volume of PBS (Component C) through a 0.22 μm pore size membrane, preparing enough for culture dilution and 500 μl per test.
- Allow the 3 mM DiOC2 (Novo et al., 2000) and 500 μM CCCP solutions (Components A and B) to come to room temperature before use.
- Wash the cells with 1 ml filtered PBS twice. Spin down at a speed of 4,000 rpm for 5 min for each wash.
- Dilute the mycobacterial culture to approximately 1 x 106 cells per ml in filtered PBS.
- Aliquot 500 μl of the bacterial suspension into a flow cytometry tube for each staining experiment to be performed. Prepare two additional tubes for a depolarized control and an unstained control.
- Add 25 μl of 500 μM CCCP (Component B) to the depolarized control sample and mix.
- Add 3 μl of 3 mM DiOC2 Component A) to each flow cytometry tube and mix (do not add stain to the unstained control sample). Incubate samples at room temperature for 30 min. Stained samples can be analyzed after 5 min, but signal intensity continues to increase until about 30 min.
- Stained bacteria can be assayed in a flow cytometer equipped with a laser emitting at 488 nm. Fluorescence is collected in the green and red channels; filters used for detecting fluorescein and the Texas Red dye, respectively, are generally suitable. The forward scatter, side scatter, and fluorescence should be collected with logarithmic signal amplification.
- Instrument adjustments are especially critical for detecting relatively small particles such as bacteria. Use the unstained control sample to locate bacterial populations in the forward and side scatter channels. Use the side scatter as the parameter for setting the acquisition trigger.
- After adjusting the flow cytometer as described above, apply the depolarized control sample. Gate on bacteria using forward versus side scatter and adjust fluorescence photomultiplier tube voltages such that the green and red MFI values are approximately equal. Do not set compensation.
- While the relative amount of red and green fluorescence intensity will vary with cell size and aggregation, the ratio of red to green fluorescence intensity can be used as a size-independent indicator of membrane potential. The data can also be processed by gating on bacteria using forward versus side scatter, and analyze gated populations with a dot plot of red versus green fluorescence reporting MFI values as linear values, not as channels.
- On a ratiometric histogram, set markers around the peaks of interest and record the mean ratio values (Figure 1). For a dot plot of red versus green fluorescence, set regions around the populations of interest and record red and green MFI values for each. To evaluate the data, divide the red population MFI by the green population MFI.
- In the flow cytometer, bacteria are identified solely on the basis of their size and stain ability.
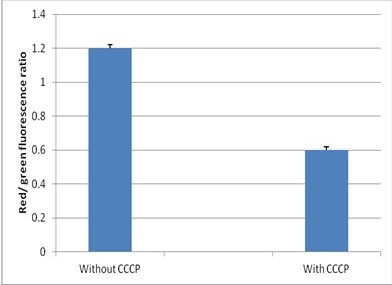
Figure 1. Detection of membrane potential in mycobacteria. Red/green ratios were calculated using population mean fluorescence intensities for mycobacteria, incubated with 3 μM DiOC2 for 30 min in either the presence or absence of 25 μM CCCP.
Notes
- Carbocyanine dyes, including DiOC2 and CCCP, are inhibitors of respiration. While these dyes do not alter assay results over the recommended staining periods, both DiOC2 and CCCP are toxic to bacterial cells and the cells will not be culturable after even brief exposure.
- The cells should be washed properly and any kind of aggregation should be avoided as it might interfere with the assay. In case any aggregates are still observed, allow the tubes to stand for a while so that the clumps can settle down. The sample can be transferred to a fresh tube.
- Solutions of the carbocyanine dye DiOC2 (3,3′-diethyloxacarbocyanine iodide) and CCCP (carbonyl cyanide 3-chlorophenylhydrazone) should be protected from light.
Recipes
- 7H9 broth supplemented with 0.1% glycerol, 0.1% TWEEN®80 and Middlebrook ADC (albumin-dextrose-catalase) enrichment
Acknowledgments
Dr. Amit Singh is a Wellcome- DBT India Alliance Intermediate Fellow. The work was supported by the Wellcome- DBT India Alliance grant, WTA01/10/355.
References
- Chawla, M., Parikh, P., Saxena, A., Munshi, M., Mehta, M., Mai, D., Srivastava, A. K., Narasimhulu, K. V., Redding, K. E., Vashi, N., Kumar, D., Steyn, A. J. and Singh, A. (2012). Mycobacterium tuberculosis WhiB4 regulates oxidative stress response to modulate survival and dissemination in vivo. Mol Microbiol 85(6): 1148-1165.
- Novo, D. J., Perlmutter, N. G., Hunt, R. H. and Shapiro, H. M. (2000). Multiparameter flow cytometric analysis of antibiotic effects on membrane potential, membrane permeability, and bacterial counts of Staphylococcus aureus and Micrococcus luteus. Antimicrob Agents Chemother 44(4): 827-834.
- Novo, D., Perlmutter, N. G., Hunt, R. H. and Shapiro, H. M. (1999). Accurate flow cytometric membrane potential measurement in bacteria using diethyloxacarbocyanine and a ratiometric technique. Cytometry 35(1): 55-63.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chawla, M. and Singh, A. (2013). Detection of Membrane Potential in Mycobacterium tuberculosis . Bio-protocol 3(11): e785. DOI: 10.21769/BioProtoc.785.
Category
Microbiology > Microbial cell biology > Cell-based analysis > Ion analysis
Cell Biology > Cell-based analysis > Flow cytometry
Cell Biology > Cell staining > Other compound
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









