- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
35S pulse Labelling of Chlamydomonas Chloroplast Proteins
Published: Vol 3, Iss 11, Jun 5, 2013 DOI: 10.21769/BioProtoc.783 Views: 10436

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Phosphopeptide Purification Protocol for the Moss Physcomitrella paten
Xiaoqin Wang and Yikun He
Jul 20, 2015 9553 Views

Indirect Immunofluorescence Assay in Chlamydomonas reinhardtii
Takashi Yamano and Hideya Fukuzawa
Jul 5, 2016 13301 Views

Tandem Purification of His6-3x FLAG Tagged Proteins for Mass Spectrometry from Arabidopsis
He Huang and Dmitri Anton Nusinow
Dec 5, 2016 13587 Views
Abstract
35S pulse labelling of proteins is used to attach a radioactive label to newly synthesized proteins, as sulfur is an element that is mainly present in proteins (Fleischmann and Rochaix 1999). Depending on your organism’s uptake mechanisms you need cysteine, methionine or sulfuric acid as a source of radioactive sulfur. This example uses Chlamydomonas cells and H235SO4 (Schwarz et al., 2012).
Keywords: 35S pulse LabellingMaterials and Reagents
- Strain of interest
- Control strains lacking the gene for the proteins of interest (as a negative control)
- Tris (Applichem, catalog number: A1379 )
- Ammonium chloride (Carl Roth, catalog number: 5470 )
- Magnesium chloride (Carl Roth, catalog number: KK36 )
- Calcium chloride (Merck, catalog number: 1023780500 )
- K2HPO4 (Applichem, catalog number: A1363 )
- KH2PO4 (Applichem, catalog number: A1364 )
- Na2EDTA (Carl Roth, catalog number: 8043 )
- ZnSO4·7H2O (Carl Roth, catalog number: T884 )
- H3BO3 (Carl Roth, catalog number: P010 )
- MnCl2·4H2O (Carl Roth, catalog number: 0 276 )
- FeSO4·7H2O (Carl Roth, catalog number: P015 )
- CoCl2·6H2O (Carl Roth, catalog number: T889 )
- CuSO4·5H2O (Carl Roth, catalog number: 8175 )
- (NH4)6Mo7O24·4H2O (Carl Roth, catalog number: 3666 )
- HEPES (Carl Roth, catalog number: HN78 )
- Tricine (Carl Roth, catalog number: 6977 )
- Methanol (Applichem, catalog number: A3493 )
- Cycloheximide (Carl Roth, catalog number: 8682 )
- 35S sulfuric acid (Hartmann Analytic, catalog number: S-RA-1 )
- Liquid nitrogen (Linde, inquire)
- Protease inhibitor cocktail (F. Hoffmann-La Roche, catalog number: 04693159001 )
- TAP-B (see Recipes)
- TAP-B/T(see Recipes)
- Hutner trace elements (see Recipes)
- Buffer A (see Recipes)
- Buffer B (see Recipes)
Equipment
- Sterile Erlenmeyer flasks (Brand KG)
- Photometer (GE Healthcare)
- Hemocytometer (Brand KG)
- Microscope (≥ 400x magnification, Leica)
- Reaction tubes (Sarstedt)
- Screw cap microreaction tubes (Sarstedt)
- Microreaction tube centrifuge with cooling capacity (Eppendorf)
- Gel dryer (Bio-Rad Laboratories)
- Whatman filter paper (GE Healthcare)
Procedure
I. Sulfur deprivation
- Grow your strain of interest and the control strains at 23 °C to the early log phase (cell density < 2 x 106 cells/ml).
- Spin down your cells (5 min, 1,000 x g, RT) in sterile 50 ml reaction tube and resuspend carefully in 10 ml TAP-B, centrifuge again (5 min, 1,000 x g, RT) and resuspend the cells in 10 ml TAP-B.
- Transfer cells to a sterile 25 ml Erlenmeyer flask (it is best to set up the medium in this flasks and put the resuspended cells back in the same flask), shake for 16 h at 23 °C in medium light (30 – 50 μmol x m-2 x s-1).
- Spin down your cells (5 min, 1,000 x g, RT) and resuspend carefully in 10 ml TAP-B/T.
- Centrifuge again (5 min, 1,000 x g, RT) and resuspend the cells in exactly 10 ml TAP-B/T, transfer cells back to 25 ml Erlenmeyer flask.
- Agitate cells for 2 h in the dark (wrap flasks with aluminium foil or cover with cardboard box).
II. Adjustment of cells to the same amount of chlorophyll/cells
- Transfer 0.5 ml of the cultures to a microreaction tube (keep the remaining culture shaking in the dark).
- Centrifuge tube (2 min, 20,000 x g, 4 °C) and discard supernatant, resuspend pellet thoroughly in 1 ml methanol.
- Centrifuge again (1 min, 20,000 x g, 4 °C) and use supernatant for chlorophyll measurement at 652 nm, dilute with methanol (prechilling not necessary) if optical density is higher than 1 (do not forget to adjust your calculation for that dilution factor).
- Calculate chlorophyll content:

- Adjust Adjust chlorophyll content with TAP-B/T to 80 μg ml-1:
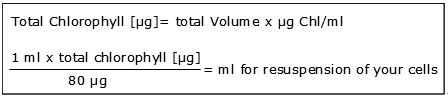
- Centrifuge cells (5 min, 1,000 x g, RT) and resuspend carefully in calculated volume of TAP-B/T to get 80 μg Chl/ml.
- If your cells are lacking chlorophyll better adjust the cell number than the chlorophyll amount, prepare a 1: 10 dilution of cultures and count the cells (hemocytometer, microscope), spin down cells and adjust cell number to 7.25 x 107 cells/ml (that corresponds to ~ 80 μg Chlorophyll/ml if using a green culture).
III. Pulse
- Add 25 μl of cycloheximide stock solution (100 μg/ml, final: 10 μg/ml) per culture to a screw cap tube (cycloheximide is for inhibition of cytosolic protein synthesis).
- Take your cultures and prepared tubes and go to fume hood in the radioactive lab (don’t inhale 35S! Release of radioactive gaseous SO2).
- Add 225 μl of each culture to the cycloheximide.
- Incubate for 10’ in rotary shaker in the dark (covered with cardboard box).
- Add 12.5 μl of 35S (H235SO4, 10 mCi/ml).
- Light pulse: incubate for 5-20 min in front of a appropriate light source and agitate cells occasionally.
- Spin down cells, discard supernatant (radioactive waste) and freeze the tube with the pellet in liquid nitrogen for at least 5 min to stop cellular activity.
IV. Cell lysis
- Thaw your cells on ice and keep them on ice from now on.
- Add 200 μl of buffer A and break the cells by pipetting up and down for ~ 1 min or sonication (three times 5 pulses at 50% output with 30’’ pauses in between).
- Remove soluble material: spin down membranes at 20,000 x g for 25 min at 4 °C, discard supernatant into radioactive waste.
- Resuspend membrane pellet with 100 μl of buffer B.
V. Protein electrophoresis
Use a protocol for Laemmli-SDS-PAGE and adjust conditions for your protein depending on its molecular weight, e.g. a 6 M urea-16% polyacrylamide-SDS-PAGE gel for separation of the photosystem II reaction center proteins D1 (encoded by the psbA gene) and D2 (encoded by the psbD gene).
VI. Coomassie staining and drying of gel
- Use a protocol for Coomassie Blue staining (to visualize size marker and lanes).
- Put gel (upside down) on plastic tray, put Whatman paper (moistened with water) on top, flip over (Figure 1).
- Place Whatman paper and gel on top of two more layers of Whatman paper, cover with plastic foil and dry in gel dryer.
VII.Autoradiography
- Place phosphor imaging screen on top of your gel and expose for 1-3 days (or use X-ray film for longer time).
- Scan screen / develop X-ray film.
- Compare bands in your strains of interest with negative controls to identify affected protein, e.g. the band for the photosystem II reaction center protein D1 (encoded by the psbA gene) is missing in the investigated mutants as in the psbA mutant FuD7 in Figure 5 of (Morais et al., 1998).
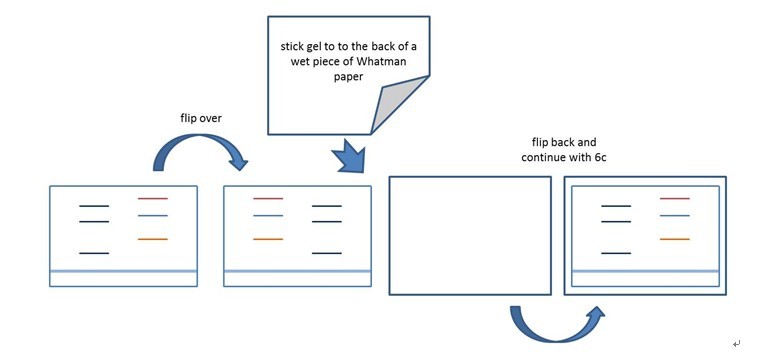
Figure 1. Scheme of step (VI-2).
Recipes
- TAP-B
20 mM Tris
7.5 mM ammonium chloride
0.805 mM magnesium chloride
0.34 mM calcium chloride
0.537 mM K2HPO4
0.463 mM KH2PO4
0.1% Hutner trace elements
Adjust to pH 7 with acetic acid
- TAP-B/T
20 mM tris
7.5 mM ammonium chloride
0.805 mM magnesium chloride
0.34 mM calcium chloride
0.537 mM K2HPO4
0.463 mM KH2PO4
Adjust to pH 7 with acetic acid
- Hutner trace elements (Hill and Kafer, 2001)
50 g Na2EDTA·2H2O
22 g ZnSO4·7H2O
11.4 g H3BO3
5 g MnCl2·4H2O
5 g FeSO4·7H2O
1.6 g CoCl2·6H2O
1.6 g CuSO4·5H2O
1.1 g (NH4)6Mo7O24·4H2O
Fill up with dH2O to 1,000 ml
- Buffer A
10 mM EDTA
10 mM HEPES (pH 7.8)
Protease inhibitor cocktail (according to the manufacturer’s instructions)
- Buffer B
10 mM EDTA
10 mM tricine (pH 7.8)
Protease inhibitor cocktail (according to the manufacturer’s instructions)
Acknowledgments
This protocol was adapted from the protocol published by Fleischmann and Rochaix (1999). The work was supported by a grant from the Deutsche Forschungsgemeinschaft to J.N. (grant number Ni390/4-2).
References
- Fleischmann, M. M. and Rochaix, J. D. (1999). Characterization of mutants with alterations of the phosphorylation site in the D2 photosystem II polypeptide of Chlamydomonas reinhardtii. Plant Physiol 119(4): 1557-1566.
- Hill T. W., Kafer E. (2001). Improved protocols for Aspergillus minimal medium: trace element and minimal medium salt stock solutions. Fungal Gen News 48: 20 -21
- Morais, F., Barber, J. and Nixon, P. J. (1998). The chloroplast-encoded alpha subunit of cytochrome b-559 is required for assembly of the photosystem two complex in both the light and the dark in Chlamydomonas reinhardtii. J Biol Chem 273(45): 29315-29320.
- Schwarz, C., Bohne, A. V., Wang, F., Cejudo, F. J. and Nickelsen, J. (2012). An intermolecular disulfide-based light switch for chloroplast psbD gene expression in Chlamydomonas reinhardtii. Plant J 72(3): 378-389.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Bohne, A., Schwarz, C. and Nickelsen, J. (2013). 35S pulse Labelling of Chlamydomonas Chloroplast Proteins. Bio-protocol 3(11): e783. DOI: 10.21769/BioProtoc.783.
Category
Plant Science > Plant biochemistry > Protein > Labeling
Biochemistry > Protein > Labeling
Biochemistry > Other compound > Chlorophyll
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









