- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Generation of Polyclonal Specific Antibodies
Published: Vol 3, Iss 11, Jun 5, 2013 DOI: 10.21769/BioProtoc.779 Views: 12476
Reviewed by: Ru Zhang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protein Immunoprecipitation Using Nicotiana benthamiana Transient Expression System
Fang Xu [...] Xin Li
Jul 5, 2015 23192 Views

Separation and Visualization of Low Abundant Ubiquitylated Forms
Ramona Schuster [...] Mafalda Escobar-Henriques
Nov 20, 2018 5515 Views

Assessing Self-interaction of Mammalian Nuclear Proteins by Co-immunoprecipitation
Claudia Cattoglio [...] Anders S. Hansen
Feb 20, 2020 9471 Views
Abstract
Generation of antibodies specific for a protein of interest is a common method in many disciplines. This protocol details the steps in production of a polyclonal antibody in rabbits using a bacterially expressed fusion protein as an antigen. The protocol is generated based on data presented in Wirschell et al.(2013).
Keywords: AntibodyMaterials and Reagents
- Topo TA cloning kit with One Shot competent cells (Life Technologies, Invitrogen™, catalog number: K457501 )
- pCR2.1 TOPO
- pET28a
- pMal-C (New England Biolabs)
- BL21 (DE3) pLysS competent cells (Stratagene, catalog number: 200132 )
- Bacto agar (BD Biosciences, catalog number: 214530 )
- Bacto Peptone Peptone (BD Biosciences, catalog number: 211677 )
- Bacto yeast extract (BD Biosciences, catalog number: 212750 )
- NaCl (Thermo Fisher Scientific, catalog number: 7647-14-5 )
- IPTG (Promega Corporation, catalog number: PR-V3953 )
- Novagen BugBuster Lysis buffer (Thermo Fisher Scientific, catalog number: 50-230-9216 )
- Urea (Thermo Fisher Scientific, catalog number: U15-500 )
- Novagen His-Bind Purification kit (Thermo Fisher Scientific, catalog number: 50-230-8606 )
- Novagen rLysozyme (EMD Millipore, catalog number: 71110-4 )
- Novagen Benzonase (EMD Millipore, catalog number: 70664-3 )
- PMSF (Sigma-Aldrich, catalog number: 78830-5G )
- Aprotinin (Sigma-Aldrich, catalog number: A1153-5MG )
- Roche Complete Protease inhibitor cocktail (F. Hoffmann-La Roche, catalog number: 04-693-124-001 )
- Takara Chaperone plasmid set (Takara Bio Company, catalog number: 3340 )
- Amylose resin (New England Biolabs, catalog number: E8021S )
- Nitrocellulose membrane (Bio-Rad Laboratories, catalog number: 162-0115 )
- PVDF (EMD Millipore, catalog number: IPVH00010 )
- I-BLOCK (Life Technologies, Invitrogen™, catalog number: T2015 )
- Tween-20 (Sigma-Aldrich, catalog number: P9416 )
- Triton X-100 (Sigma-Aldrich, catalog number: T8787 )
- Sodium azide (Sigma-Aldrich, catalog number: S2002 )
- Glycine (Sigma-Aldrich, catalog number: G8898 )
- Tris (Sigma-Aldrich, catalog number: T1503 )
- Sodium bicarbonate (Sigma-Aldrich, catalog number: S5761 )
- AminoLink Immobilization kit (Pierce Antibodies, catalog number: 44894 )
- Hydrochloric acid (HCl) (Thermo Fisher Scientific, catalog number: A144-212 )
- Sodium acetate (Sigma-Aldrich, catalog number: S2889 )
- Ethanol by Acros(Thermo Fisher Scientific, catalog number: 61509-0020 )
- Methanol (Thermo Fisher Scientific, catalog number: BP1105-4 )
- Na2HPO4
- KH2PO4
- KCl
- NaCl
- CNBr-activated sepharose (Sigma-Aldrich, catalog number: C9142 )
- 3x PBST (see Recipes)
- Glycine elution buffer (see Recipes)
- Low pH wash buffer (see Recipes)
- High pH wash buffer (see Recipes)
- Coupling buffer (see Recipes)
Equipment
- Nickel-chromatography column (Thermo Fisher Scientific, catalog number: 50-230-8606 )
Procedure
- Preparation of expression construct:
- cDNA sequences encoding amino acids 155-243 of Chlamydomonas DRC1 were amplified by PCR and cloned into the pCR2.1 TOPO cloning vector to generate plasmid pMW199.1.
- The insert from pMW199.1 was excised with EcoR1 and subcloned into the EcoR1 site of pET28a to generate plasmid pMW219.15, which was sequenced to confirm orientation. The expressed protein sequence is shown in Figure 1.

Figure 1. The DRC-His fusion protein. The predicted size of the expressed fusion protein is ~20 kDa. In bold is the sequence encoded by the pET28A vector including the 6-His tag (underlined) followed by the sequences encoded by the EcoR1 insert from pMW199.1 containing 6 amino acids from the pCR2.1 cloning vector and DRC1 sequences (italics).
- cDNA sequences encoding amino acids 155-243 of Chlamydomonas DRC1 were amplified by PCR and cloned into the pCR2.1 TOPO cloning vector to generate plasmid pMW199.1.
- Production of the His-tagged DRC1 fusion protein: 1 ng of pMW219.15 was transformed into 50 μl of BL21 (DE3) pLysS cells from Stratagene by electroporation according to manufacturer’s recommendations.
- Single colonies were selected and tested for expression.
- Cells were grown overnight in 3 ml LB cultures at 37 °C.
- The next day, cultures were diluted to A600 = 0.1 and grown until A600 = 0.3 - 0.4.
- Expression of the His-DRC1 fusion was induced with 100 mM IPTG for 2-3 h at 37 °C.
- Cells were lysed and both soluble and insoluble fractions were analyzed by 15% SDS-PAGE for induction of the fusion protein.
- The His-DRC1 fusion protein was completely insoluble and found in inclusion bodies.
- Cells were grown overnight in 3 ml LB cultures at 37 °C.
- Scaled-up expression:
- Cells were grown in 50 ml overnight LB cultures at 37 °C.
- The next day, cells were diluted into 1.5 L LB to an A600 = 0.1 and grown to A600 = 0.3.
- Expression of the His-DRC1 fusion protein was induced with 100 mM IPTG for 2 h.
- Cells were grown in 50 ml overnight LB cultures at 37 °C.
- Single colonies were selected and tested for expression.
- Lysis of bacterial cells and purification of inclusion bodies:
- Cells were pelleted and resuspended in 30 ml (5 ml/gram of cell pellet) BugBuster supplemented with 10 U/ml rLysozyme, 10 U/ml Benzonase, 1 mM PMSF, 2.5 U/ml Aprotinin (Alternatively, use Roche complete protease inhibitor cocktail per manufacturers instructions). Incubate for 30 min at room temperature on a platform or rotisserie shaker.
- Add Triton-x-100 to 0.1% and incubate 10 min.
- Centrifuge at 5,000 x g for 15 min at 4 °C.
- Save supernatant (S1) for gel analyses. Resuspend the pellet in 0.5 volumes (15 ml) of Bugbuster diluted 1:10 with water (0.1x Bugbuster). Vortex to resuspend the pellet. Centrifuge 16,000 x g for 15 min at 4 °C.
- Save supernatant (S2) for gel analyses. Resuspend the pellet in 15 ml of 0.1x Bugbuster. Vortex to resuspend the pellet. Centrifuge at 16,000 x g for 15 min at 4 °C.
- Save supernatant (S3) for gel analyses. Resuspend the pellet in 1x binding buffer + 6 M urea. Save an aliquot for gel analyses (IB).
- Cells were pelleted and resuspended in 30 ml (5 ml/gram of cell pellet) BugBuster supplemented with 10 U/ml rLysozyme, 10 U/ml Benzonase, 1 mM PMSF, 2.5 U/ml Aprotinin (Alternatively, use Roche complete protease inhibitor cocktail per manufacturers instructions). Incubate for 30 min at room temperature on a platform or rotisserie shaker.
- Purification of His-DRC1 using Novagen His-Bind purification kit:
- Add 2.0 ml of resin to a column (20 mg/2.5 ml binding capacity) and let drain.
- Wash resin with 3 volumes distilled water.
- Wash resin with 5 volumes of 1x charge buffer (50 mM NiSO4).
- Wash with 3 volumes of 1x binding buffer + 6 M urea.
- Connect column to a peristaltic pump with a flow rate set at ~5 ml/h (0.08 ml/min). Load the solubilized inclusion bodies containing the His-DRC1 fusion protein onto the column and let run through the column. Save flow through (FT) for gel analyses.
- Wash column with 10 column volumes of 1x binding buffer + 6 M urea.
- Wash column with 6 column volumes of 1x wash buffer + 6 M urea + 20 mM Imidizole.
- Flush out tubing briefly and reconnect to column and peristaltic pump.
- Elute fusion protein in 6 column volumes of 1x elution buffer + 6 M urea and collect in 10x 300 μl fractions.
- Fix 50 μl of each fraction for gel analyses.
- Run all samples (saved supernatants, purified inclusion bodies, flowthrough, washes and column fractions) on 12% acrylamide SDS-PAGE gel (Figure 2).
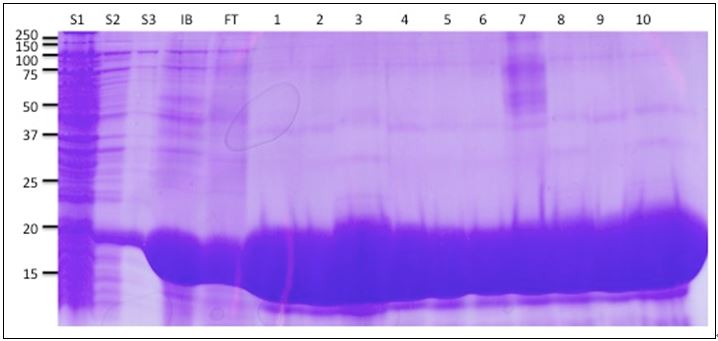
Figure 2. Coomassie stained gel of DRC1-His purification. S1, S2 and S3 are the supernatants from the three washes of the insoluble cell pellet. IB is the inclusion body pellet. FT is the flow through from the Nickel column. IB is the inclusion body pellet. 1-10 are the fractions collected from the column.
- Add 2.0 ml of resin to a column (20 mg/2.5 ml binding capacity) and let drain.
- Immunization of rabbits for polyclonal antibody production: The antibody against DRC1 was generated using the standard protocol offered by Spring Valley Laboratory (Woodbine, MD).
- First, we requested pre-immune sera from 10 rabbits. These were tested on western blots for endogenous reactivity to axonemal proteins. 2 rabbits were selected that showed minimal to no reactive bands on western blots.
- 4.2 mg of column purified His-DRC1 fusion protein was sent to Spring Valley Laboratories to be used as antigen for immunizations. The fusion protein was in the elution buffer used to elute the fusion protein from the Nickel-chromatography column.
- The following protocol was followed for immunization and bleeding:
- Day 0: Pre-immunization test bleed
- Day 0: Primary immunization
- Day 21: Immunogen boost
- Day 42: Immunogen boost
- Day 52: Bleed
- Day 63: Immunogen boost
- Day 73: Bleed or exsanguination
Optional: monthly protocol extension
Day 1: Immune boost
Day 12: bleed
Day 27: bleed - For our DRC1 antibody production, we opted to exsanguinate after the day 73 production bleed. Approximately 1 mg of total protein is used per rabbit for the immunizations.
- Day 0: Pre-immunization test bleed
- First, we requested pre-immune sera from 10 rabbits. These were tested on western blots for endogenous reactivity to axonemal proteins. 2 rabbits were selected that showed minimal to no reactive bands on western blots.
- Purification of DRC1-specific antibodies: Whole serum was tested by western blot on isolated axonemes from wild-type cells and the drc1-mutant to verify specificity of the antibody produced in rabbits. The sera was used at a 1:10,000 dilution for western blots or was further affinity purified as follows:
- MBP-DRC1 fusion protein:
- An MBP-DRC1 construct was prepared by inserting the EcoR1 insert from pMW199.1 into the EcoR1-digested pMal-C vector. Isolated clones were sequence verified.
- The MBP-DRC1 fusion protein was expressed in the presence of chaperone proteins to increase the solubility of the fusion protein (Takara Chaperone plasmid set).
- Soluble MBP-DRC1 fusion protein was purified on an amylose resin according to manufacturers instructions, run on 10% SDS-PAGE at 200 V for 1 h and transferred to nitrocellulose or PVDF membrane. The region of the membrane containing the fusion protein was cut out and used to purify DRC1-specific antibodies.
- An MBP-DRC1 construct was prepared by inserting the EcoR1 insert from pMW199.1 into the EcoR1-digested pMal-C vector. Isolated clones were sequence verified.
- Blot affinity purification:
- The MBP-DRC1 fusion protein was run on SDS-PAGE and transferred to two sheets of PVDF membrane. The region of the membranes containing the fusion protein were cut out and used to purify DRC1-specific antibodies.
- The MBP-DRC1 membrane was washed in 1x PBS, and then blocked with 5 ml of 0.2% I-BLOCK, 0.1% Tween-20 in PBS for 30 minutes.
- The strip was incubated overnight at room temperature with a 1:1,000 dilution of DRC1 sera in the blocking solution plus 0.05% azide to specifically bind DRC1 antibodies to the membrane.
- The membrane was washed 3x in PBS-Tween, and the bound DRC1 antibodies were eluted in 2 ml of glycine elution buffer for 3 min on ice.
- The glycine eluate was collected and neutralized with 1/10th volume of 1 M Tris pH 8.1.
- The elution step was repeated two more times and the three elutes were combined, normal goat serum was added to 10%, and then the eluted antibody was dialyzed overnight against 1x PBS at 4 °C.
- The eluted, dialyzed antibody was supplemented with 0.1% azide and stored at 4 °C.
- The MBP-DRC1 fusion protein was run on SDS-PAGE and transferred to two sheets of PVDF membrane. The region of the membranes containing the fusion protein were cut out and used to purify DRC1-specific antibodies.
- Column affinity purification:
- The MBP-DRC1 fusion protein was purified as indicated above and peak fractions (~10 mg) dialyzed overnight at 4 °C into coupling buffer.
- Next day: Prepare the CNBr-activated sepharose. The binding capacity is 13-20 mg/ml. Weigh out 0.3 grams for ~10 mg of MBP-DRC1 fusion protein and rehydrate / wash it with 1 mM HCl.
- Wash the column with water, then coupling buffer.
- Bind the MBP-DRC1 fusion to the CNBr-activated sepharose column for 2.5 h at room temperature using a rabbit pump to recirculate the protein back onto the column.
- Wash the column with alternating low pH wash buffer and high pH wash buffer. The column was stored in 20% ethanol until ready to use for affinity purification.
- Wash the MBP-DRC1 column with PBS.
- Cap the bottom of the column and add 2 ml of DRC1 sera and 500 μl of PBS.
- Mix and let the resin re-settle in the column. Incubate for 1 h.
- Collect the flow through.
- Wash the column with PBS.
- Elute antibody with 6 ml of 0.1 M Glycine pH 2.87.
- Collect 900 μl fractions into Eppendorf tubes with 100 μl of 1 M Tris pH 8.0.
- Pool the peak fractions and dialyze overnight into PBS.
- Collect the dialyzed antibodies and add sodium azide as a preservative.
- The MBP-DRC1 fusion protein was purified as indicated above and peak fractions (~10 mg) dialyzed overnight at 4 °C into coupling buffer.
- Pre-absorption to methanol-fixed drc1-mutant cells:
- The pf3 mutant cells were grown in standard Chlamydomonas media (Harris, 2009)
- Cells were harvested, washed and resuspended in ~50 ml of 100% methanol repeatedly until the supernatant was clear and the pellet was white. The final pellet of extracted cells was split into 5 aliquots.
- 50-100 microliters of antisera was diluted 1:10 into PBS plus 0.1% azide and then incubated overnight at 4 °C with the first aliquot of extracted cells.
- The supernatant was removed, a sample saved, and then the supernatant was incubated overnight with the 2nd aliquot of extracted cells. The process was repeated three more times.
- The original 1:10 dilution of antisera was diluted 1:1,000, and the supernatants from each absorption step were diluted 1:100 and 1:1,000. All samples were tested on western blots of wild-type and pf3 mutant axonemes. Aliquots with minimal background, but strong signals, were combined and used at the appropriate dilution (Wirschell et al., 2013).
- The pf3 mutant cells were grown in standard Chlamydomonas media (Harris, 2009)
- MBP-DRC1 fusion protein:
Recipes
Note: All buffers can be generated using the concentrated stock solutions provided with the referenced kits and by standard molarity calculations.
- 3x PBST
9.6 mM Na2HPO4
1.5 mM KH2PO4
3.9 mM KCl
405 mM NaCl
0.15% Tween-20 - Glycine elution buffer
0.1 M glycine-HCl
0.5 M NaCl
0.05% Tween (pH 2.5) - Low pH wash buffer
0.1 M sodium acetate (pH 3.85)
0.5 M NaCl - High pH wash buffer
0.1 M Tris (pH 9)
0.5 M NaCl - Coupling buffer
0.1 M NaHCO3 (pH 8.0)
0.5 M NaCl
References
- Harris, E. (2009). Chlamydomonas in the Laboratory. The Chlamydomonas Sourcebook. E. Harris. Kidlington, Oxford, Academic Press. 1: 241-301.
- Wirschell, M., Olbrich, H., Werner, C., Tritschler, D., Bower, R., Sale, W. S., Loges, N. T., Pennekamp, P., Lindberg, S., Stenram, U., Carlen, B., Horak, E., Kohler, G., Nurnberg, P., Nurnberg, G., Porter, M. E. and Omran, H. (2013). The nexin-dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat Genet 45(3): 262-268.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wirschell, M. and Porter, M. E. (2013). Generation of Polyclonal Specific Antibodies. Bio-protocol 3(11): e779. DOI: 10.21769/BioProtoc.779.
Category
Immunology > Antibody analysis > Antibody-antigen interaction
Biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Immunodetection > Immunoprecipitation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









