- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Packaging of Retroviral RNA into Viral Particles Analyzed by Quantitative Reverse Transcriptase-PCR
Published: Vol 3, Iss 8, Apr 20, 2013 DOI: 10.21769/BioProtoc.684 Views: 17244
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A SYBR Green-based Real Time RT-PCR Assay for Detection of the Emerging H7N9 Virus
Zheng Zhu and Lunbiao Cui
Jun 20, 2014 10104 Views

Infectious Virus Yield Assay for Hepatitis E Virus
Yannick Debing [...] Johan Neyts
Aug 5, 2014 11477 Views

Quantification of HIV RNA and Human Herpesvirus DNA in Seminal Plasma
Milenka V. Vargas-Meneses [...] Sara Gianella
May 5, 2015 11129 Views
Abstract
Formation of viral particles and packaging of genomic retroviral RNA into these particles are important steps in the late phase of the viral replication cycle. The efficiency of the incorporation of viral or cellular RNAs into viral particles can be studied using a quantitative Reverse Transcriptase-PCR (RT-qPCR)-based approach. After isolation of cytoplasmic RNA from either infected or transfected cells and extraction of virus particle-associated RNA, specific RNA levels present in both fractions are determined. The ratio of virion-associated and cytoplasmic RNA defines the encapsidation efficiency (Brandt et al., 2007; Blissenbach et al., 2010; Grewe et al., 2012).
Keywords: HIVMaterials and Reagents
I. Cell harvesting and ultracentrifugation
- Dulbecco’s Modified Eagle Medium (DMEM) (Life Technologies, Gibco®, catalog number: 41965-039 )
- Fetal calf serum (FBS) (Life Technologies, Gibco®, catalog number: 10108-165 )
- Phosphate buffered saline (PBS) (Life Technologies, Gibco®, catalog number: 14200-067 )
- Trypsin (Biochrom)
- Penicillin/Streptomycin (Life Technologies, Gibco®)
- 0.45 μm filter (Sarstedt)
- Sucrose (Biomol)
- Ultracentrifugation tubes (17 ml) (Herolab)
II. Cell fractionation and RNA isolation
- Dithiothreitol (DTT) solution (1 M) (AppliChem GmbH, catalog number: A3668,0050 )
- RiboLock RNase Inhibitor (40 U/μl) (Thermo Fisher Scientific, catalog number: E00384 )
- Ethanol >99.8% (Sigma-Aldrich)
- QIAshredder Spin Columns (QIAGEN, catalog number: 79654 )
- RNeasy Kit (QIAGEN, catalog number: 74106 )
- QIAmp Viral RNA Kit (QIAGEN, catalog number: 52906 )
- TURBO DNA-free Kit (Life Technologies, Ambion®, catalog number: AM1907 )
- Nuclease-free water (B Braun)
- Quant-iT RNA Assay Kit (Life Technologies, InvitrogenTM, catalog number: Q-33140 )
III. RT-qPCR
- QuantiTect Probe RT-PCR Kit (includes Reverse Transcriptase) (QIAGEN, catalog number: 204443 )
- SYBR Green (Life Technologies, InvitrogenTM)
- Primers sense and antisense (Biomers)
- Isolated RNA
- TOPO TA Cloning Kit (Life Technologies, InvitrogenTM, catalog number: 45-0640 )
- AmpliScribe-T7/Sp6 High Yield Transcription Kit (Epicentre, catalog number: AS2607 )
- RNA standards
IV. Other materials
- HIV-1 DIY p24 sandwich ELISA Kit 2 (Aalto Bio Reagents)
Equipment
- Heraeus Multifuge X3R Centrifuge (Thermo Fisher Scientific)
- Centrifuge 5417R (Eppendorf)
- Sorvall WX Ultra 100 Ultracentrifuge (Thermo Fisher Scientific)
- Qubit Fluorometer (Life Technologies, InvitrogenTM, catalog number: Q32857 )
- Rotor-Gene Q (QIAGEN, catalog number: 9001620 )
- Strip tubes 0.1 ml (QIAGEN)
- 15 ml tubes (Sarstedt)
- 1.5 ml tubes (Sarstedt)
Procedure
Note: The procedure was established for cell numbers obtained from small 25 cm2 cell culture flasks. Initially, 1.5 million adherent cells were seeded. After transfection/infection and subsequent cultivation the cells reached 80-100% confluence at day of cell harvest (three days later).
- Concentration of viral particles
- Centrifuge supernatants of adherent, transfected or infected cells at 300 x g and 4 °C for 5 min to pellet all remaining cellular material. When suspension cells are used, pellet the cells by this centrifugation step. For 25 cm2 flasks 5 ml medium is used during transfection and cultivation. In general, a larger volume of medium can be used as long as the capacity of the ultracentrifugation tubes (see I-8 in materials and step 1-d of the procedure) including the sucrose solution is not exceeded.
- Add fresh medium to the transfected or infected cells and place them in the incubator until cytoplasmic RNA isolation.
- Filter cell-free supernatants through a 0.45 μm filter.
- Ultracentrifuge pre-cleared supernatants through a 30% sucrose cushion. Add 2.5 ml sucrose solution to the tube and carefully overlay with the 5 ml supernatant without disrupting the sucrose pellet. The tubes have to be filled up with serum-free DMEM until 0.5-1 cm below the edge of the tube. This is important to stabilize the tubes during ultracentrifugation. The final volume is determined by precise balancing the tubes in buckets which are inserted opposite each other in the same rotor. Centrifuge at 150,000 x g for 2 h at 4 °C.
- Centrifuge supernatants of adherent, transfected or infected cells at 300 x g and 4 °C for 5 min to pellet all remaining cellular material. When suspension cells are used, pellet the cells by this centrifugation step. For 25 cm2 flasks 5 ml medium is used during transfection and cultivation. In general, a larger volume of medium can be used as long as the capacity of the ultracentrifugation tubes (see I-8 in materials and step 1-d of the procedure) including the sucrose solution is not exceeded.
- Harvest cells
- Adherent cells: Discard the freshly added cellular supernatant, wash cells once with PBS (1x; RT) and detach cells with trypsin. Stop trypsin treatment by resuspending cells in serum-containing medium. Pellet cells at 300 x g and 4 °C for 10 min and resuspend them in cold PBS (4 °C).
- Suspension cells: Centrifuge cells at 300 x g and 4 °C for 10 min, discard freshly added cellular supernatant and resuspend cells in cold PBS (4 °C).
- Pellet cells at 300 x g and 4 °C for 5 min and discard the supernatant. Remove all remaining PBS and loosen the cells by flipping the tube a few times.
- During centrifugation steps: Prepare working solutions of RLN and RLT buffers. Prepare 600 μl aliquots of RLT buffer in 1.5 ml tubes. Store both buffers on ice.
- Adherent cells: Discard the freshly added cellular supernatant, wash cells once with PBS (1x; RT) and detach cells with trypsin. Stop trypsin treatment by resuspending cells in serum-containing medium. Pellet cells at 300 x g and 4 °C for 10 min and resuspend them in cold PBS (4 °C).
- Cell fractionation
- Resuspend cells carefully in 175 μl ice cold RLN buffer by shortly pipetting them up and down with a wide bored 1,000 μl pipette. Extensive pipetting may result in total cell lysis and should be prevented.
- Incubate exactly 5 min on ice to lyse the plasma membrane.
- Pellet nuclei by centrifugation at 300 x g for 2 min at 4 °C in a pre-cooled centrifuge.
- Transfer the supernatant representing the cytoplasmic fraction to new 1.5 ml tubes with a 200 μl pipette. Be sure not to contaminate the supernatant with the nuclear pellet.
- Centrifuge the cytoplasmic fraction at 13,000 x g at 4 °C for 3 min in a pre-cooled centrifuge to pellet all remaining nuclear contaminants.
- Transfer supernatant to already prepared 600 μl RLT aliquots (step 2-c) and store on ice.
- Optional: Extraction of the nuclear fraction
- Resuspend nuclei pellet in 900 μl cold PBS (4 °C) and centrifuge for 5 min at 300 x g and 4 °C in a pre-cooled centrifuge.
- Discard PBS, add 600 μl icecold RLT buffer and mix by vortexing. Cell debris will not resolve completely in the RLT buffer.
- Load the solution containing the debris onto QIAshredder Spin Columns and centrifuge 2 min at 13,000 x g. Collect the flow through as nuclear fraction.
- Resuspend nuclei pellet in 900 μl cold PBS (4 °C) and centrifuge for 5 min at 300 x g and 4 °C in a pre-cooled centrifuge.
- Resuspend cells carefully in 175 μl ice cold RLN buffer by shortly pipetting them up and down with a wide bored 1,000 μl pipette. Extensive pipetting may result in total cell lysis and should be prevented.
- Cytoplasmic/Nuclear RNA isolation
- Vortex nuclear or cytoplasmic fractions in RLT and spin for 5 sec at 13,000 x g and RT.
- Add 430 μl ethanol (> 99.8%) and mix by pipetting up and down.
- Isolate RNA with RNeasy Mini Kit according to manufacturer’s instructions.
- Elute RNA with 47 μl RNase-free H2O (this will result in an elution volume of 45 μl).
- Perform DNase digestion with the TURBO DNase-free Kit as follows: Add 5 μl 10x TURBO-DNase buffer and 1 μl or 2 μl TURBO DNase to cytoplasmic or nuclear eluates, respectively. Mix gently and incubate 2-4 h at 37 °C.
- Inactivate DNase by adding 5 μl inactivation reagent (included in TURBO DNA-free Kit) per 1 μl DNase used, incubate for 5 min at RT while keeping the reagent resuspended.
- Centrifuge 1 min at 11,000 rpm and RT and transfer the supernatant into a fresh tube.
- Measure RNA concentration using an RNA-specific dye (e.g. Quant-iT RNA Assay Kit), adjust RNA concentrations with nuclease-free water to e.g. 0.05-0.5 μg/μl and store samples at -80 °C.
- Vortex nuclear or cytoplasmic fractions in RLT and spin for 5 sec at 13,000 x g and RT.
- Isolation of particle-associated RNA
- Carefully remove the supernatant from ultracentrifuged samples from step 1-d and dry the tube walls with a clean wipe.
- Resuspend (invisible) pellet in 150 μl PBS by pipetting up and down (Optional: Shake tubes 1 h on ice in advance. This step allows a 1 h interruption of the procedure without any loss of assay performance).
- Take 10 μl of concentrated, resuspended particles and store at -20 °C for p24-specific ELISA (HIV-1 DIY p24 sandwich ELISA Kit Protocol 2).
Note: For analyzing the encapsidation efficiency virion-associated RNA copy numbers need to be normalized to the amount of particles in the supernatant. (Reduced/increased levels of particle-associated RNA can sometimes be attributed to reduced/increased particle levels.) Therefore, determination of p24 levels in the supernatant is important. - Add 560 μl AVL buffer containing carrier RNA (both included in QIAmp Viral RNA Kit) to the samples, vortex and incubate 10 min at RT (crystals appear in the AVL buffer containing carrier RNA when stored at 4 °C. Resolve those precipitations by vigorous shaking for max. 2 min at 50 °C and let the buffer cool down to RT before usage).
- Add 560 μl ethanol (> 99.8%) to the samples and vortex vigorously for 15 sec.
- Isolate RNA using QIAmp Viral RNA Kit according to the manufacturer’s instructions.
- Elute virus-associated RNA with 47 μl RNase-free H2O.
- Perform DNase digestion as described in steps 4-e~4-g. An amount of 1 μl DNase is sufficient for particle-RNA samples.
- Store samples at -80 °C until further use. Measurement of RNA concentration is not necessary in this case, because the total RNA amount is low and the samples contain unspecific carrier RNA (present in the AVL buffer).
- Carefully remove the supernatant from ultracentrifuged samples from step 1-d and dry the tube walls with a clean wipe.
- RT-qPCR
- Set up RT-qPCR reaction using QuantiTect Probe RT-PCR Kit as follows:
Isolated RNA0.5 μg cytoplasmic or 0.05 μg nuclear RNA; 5 μl particle-associated RNA; 5 μl RNA standards ranging from 2 x 101-1 x 109 RNA copies/μlPCR mix (2x, contains Taq, dNTPs) 10 μl Reverse Transcriptase (RT) enzyme 0.2 μl Primer sense (2.5 μM) 1 μl Primer antisense (2.5 μM) 1 μl SYBR Green (0.1x) 1 μl
Add nuclease-free water to 20 μl.
Prepare additional samples omitting the RT enzyme in order to detect amplification of (genomic or transfected) DNA sequences.
Primers used for HIV-1 specific RT-qPCR are used according to the Amplicor HIV-1 Monitor Test (Michael et al., 1999).
Sense: SK145s (5’-AGT GGG GGG ACA TCA AGC AGC CAT GCA AAT-3’)
Antisense: SKCC1Bas (5’-TAC TAG TAG TTC CTG CTA TGT CAC TTC C-3’) - RNA standards identical in sequence to the detected HIV RNA segment for absolute quantification of cytoplasmic and particle-associated RNA copy numbers were produced with the AmpliScribe-T7/Sp6 High Yield Transcription Kit (Epicentre). The amplicon obtained with primer pair SK145s/SKCC1Bas after RT-(q)PCR was subcloned into the pCRII-TOPO plasmid with the TOPO TA Cloning Kit. An amplicon containing either the T7 or the SP6 promoter was used as template for the in vitro transcription reaction according to the manufacturer’s instructions. Template DNA was removed by digestion with the MBU DNase from the in vitro transcription kit and in addition by two treatments with the TURBO DNA-free kit. RNA concentrations were measured and adjusted to 2 x 101 to 1 x 109 RNA copies per μl.
- Using QuantiTect Probe RT-PCR Kit allows the RT reaction and PCR amplification to happen in one tube without interruption.
RT-qPCR is performed in a Rotor-Gene Q system with the following settings:
Steps( iii-vi) should be repeated in a total of 45 cycles.RT step 50 °C 20 min Initial denaturation 95 °C 15 min Denaturation 95 °C 10 sec Annealing 65 °C 60 sec Elongation and fluorescence 72 °C 30 sec Detection 1 Fluorescence detection 2 78 °C 15 sec - After the last PCR cycle the following melting curve measurement is performed:
Ranging from 50 °C to 99 °C with 1 °C/step, 90 sec equilibration for the first step after that 4 s/step. Since amplification is monitored by SYBRGreen in this RT-qPCR, melting curve analyses are important to verify amplification of specific DNA fragments. Furthermore, regular agarose gel electrophoresis analyses should be included to verify amplification of DNA fragments of the expected size. - Analyze RNA copy numbers with Rotor-Gene Q software using values for fluorescence measurement 2 only. In contrast to measurement 1 fluorescence signals from primer dimers should be strongly reduced at 78 °C. Please note that the exact temperature for the second fluorescence measurement may vary between different Rotor-Gene devices and therefore needs to be optimized.
- Note: The primers and settings described here are designed for the detection of HIV-1 unspliced, genomic RNA. To analyze RNA from other retroviruses the RT-qPCR protocol has to be modified.
- Set up RT-qPCR reaction using QuantiTect Probe RT-PCR Kit as follows:
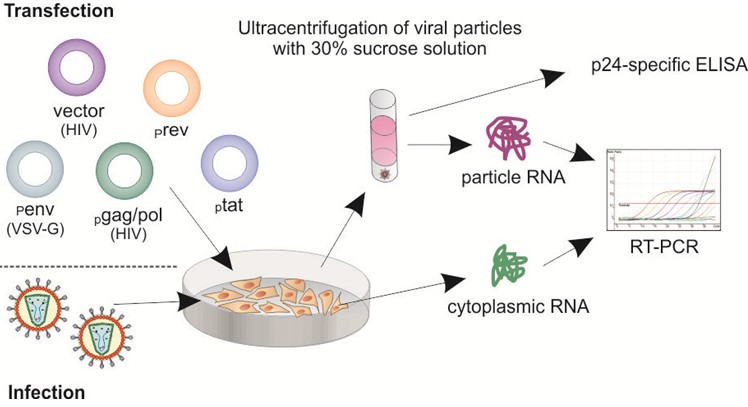
Figure 1. Schematic diagram of encapsidation assay workflow. Particle-containing supernatants from infected or transfected cells were ultracentrifuged through a 30% sucrose cushion. RNA isolated from concentrated particles and cytoplasm of the infected/transfected cells was analyzed by RT-qPCR. Encapsidation efficiency was determined as the ratio between HIV-1 copy numbers/ml supernatant and HIV-1 copy numbers/μg cytoplasmic RNA. Depending on the experimental design quantification of p24 levels in the concentrated-particle samples is important to normalize particle-associated RNA levels to the amount of p24/Gag particles in the supernatants.
Recipes
- Cell culture medium
DMEM
10% (v/v) FCS
1% (v/v) Penicillin/Streptomycin - RLN buffer
50 mM Tris-Cl (pH 8.0)
140 mM NaCl
1.5 mM MgCl2
0.5% (v/v) Nonidet P-40
Filter through a 0.2 μm filter.
Before use add 1,000 U/ml RNase inhibitor and 1 mM DTT. - RLT buffer (included in the RNeasy Mini Kit)
Before use add 1% (v/v) beta-Mercaptoethanol - AVL buffer (included in the QIAamp Viral RNA Kit)
Add 1 ml of AVL buffer to one tube of lyophilized carrier RNA (included in the QIAamp Viral RNA Kit), mix thoroughly and transfer the solution to the AVL buffer bottle. After mixing prepare aliquots and store them at 4 °C
Acknowledgments
This protocol was adapted from our papers: Brandt et al. (2007); Blissenbach et al. (2010); and Grewe et al. (2012). This work was funded by a grant from the German Research Foundation (DFG) to Klaus Überla (Ue45/11-1). Bianca Hoffmann is and Bastian Grewe was supported by a fellowship from the DFG graduate school (GRK 1045). Beside Bianca Hoffmann and Bastian Grewe, Inga Ohs, Maik Blissenbach, Sabine Brandt, Bettina Tippler, Thomas Grunwald, Klaus Überla, Rebecca Konietzny, and Klaus Sure were part of the team which established the methods described.
References
- Brandt, S., Blissenbach, M., Grewe, B., Konietzny, R., Grunwald, T. and Uberla, K. (2007). Rev proteins of human and simian immunodeficiency virus enhance RNA encapsidation. PLoS Pathog 3(4): e54.
- Blissenbach, M., Grewe, B., Hoffmann, B., Brandt, S. and Uberla, K. (2010). Nuclear RNA export and packaging functions of HIV-1 Rev revisited. J Virol 84(13): 6598-6604.
- Grewe, B., Hoffmann, B., Ohs, I., Blissenbach, M., Brandt, S., Tippler, B., Grunwald, T. and Uberla, K. (2012). Cytoplasmic utilization of human immunodeficiency virus type 1 genomic RNA is not dependent on a nuclear interaction with gag. J Virol 86(6): 2990-3002.
- Michael, N. L., Herman, S. A., Kwok, S., Dreyer, K., Wang, J., Christopherson, C., Spadoro, J. P., Young, K. K., Polonis, V., McCutchan, F. E., Carr, J., Mascola, J. R., Jagodzinski, L. L. and Robb, M. L. (1999). Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J Clin Microbiol 37(8): 2557-2563.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hoffmann, B. and Grewe, B. (2013). Packaging of Retroviral RNA into Viral Particles Analyzed by Quantitative Reverse Transcriptase-PCR. Bio-protocol 3(8): e684. DOI: 10.21769/BioProtoc.684.
Category
Microbiology > Microbial genetics > RNA > qRT-PCR
Molecular Biology > RNA > qRT-PCR
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









