- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
p65 Chromatin Immunoprecipitation Protocol
Published: Vol 3, Iss 8, Apr 20, 2013 DOI: 10.21769/BioProtoc.683 Views: 12598
Reviewed by: Lin Fang

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Identification of Proteins Interacting with Genomic Regions of Interest in vivo Using Engineered DNA-binding Molecule-mediated Chromatin Immunoprecipitation (enChIP)
Toshitsugu Fujita and Hodaka Fujii
May 20, 2014 15467 Views

RNA Chromatin Immunoprecipitation (RNA-ChIP) in Caenorhabditis elegans
Germano Cecere and Alla Grishok
Dec 20, 2014 15660 Views

Chromatin Immunoprecipitation (ChIP) Assay for Detecting Direct and Indirect Protein – DNA Interactions in Magnaporthe oryzae
Gang Li [...] Richard A. Wilson
Nov 5, 2015 15991 Views
Abstract
Chromatin Immunoprecipitation (ChIP) is an important procedure that allows you to verify if a certain protein is physically located at a regulatory region. This information, taken together with other procedures such as luciferase assays and EMSAs, will give definitive proof that the query protein is involved in the transcription of a protein. This procedure for p65 ChIP can be adapted to investigate other proteins; just a change of the antibody will suffice.
The transcription factor known as NF-κB is a homo- or hetero-dimer consisting of members of the Rel/NFKB family. The most abundant NF-κB complexes are made of two different proteins, p65 (Rel-A) and p50 (NFKB1). The NF-κB complex is initially inhibited by IκB by direct binding, thus trapping NF-κB in the cytoplasm. After a stimulatory signal, IκB kinase (IKK) phosphorylates IκB, allowing IκB to undergo proteasome-mediated degradation. The degradation of IκB and phosphorylation of p65 by multiple kinases activates NF-κB, allowing it to transport to the nucleus and cause the transcriptional activation of many of its target genes containing κB sites (consensus sequence: gggRNNYYcc, R = purine Y = pyrimidine), such as PUMA, IL-6, and TNF.
Materials and Reagents
- HCT116 cell line
- Trypsin (0.05%) (Life Technologies, catalog number: 25300-054 )
- DMSO
- PBS
- Formaldehyde (J.T. Baker, catalog number: 2106-01 )
- 1 M Glycine
- Liquid nitrogen or a dry ice/100% ethanol
- NP-40
- Protease inhibitor cocktail tablet (F. Hoffmann-La Roche, catalog number: 04-693159-001 )
- p65 antibody (Santa Cruz, catalog number: sc-109 )
- Chromatin Immunoprecipitation Assay Kit (EMD Millipore, catalog number: 17-295) online at: http://www.millipore.com/coa.nsf/a73664f9f981af8c852569b9005b4eee/20c0dc520d2b30f0852573be007ffdae/$FILE/17-295-DAM1411265.pdf
- Protein A/G PLUS-agarose (has been preblocked with BSA) (Santa Cruz, catalog number: sc-2003 )
- Phenol
- Chloroform
- Glycogen
- 70% ethanol
- SDS
- Triton X-100
- EDTA
- Tris-HCl
- NaCl
- NaHCO3
- EBC buffer (see Recipes)
- ChIP dilution buffer (see Recipes)
- 10x protease inhibitor solution (see Recipes)
- Low salt immune complex wash buffer (see Recipes)
- High salt immune complex wash buffer (see Recipes)
- LiCl immune complex wash buffer (see Recipes)
- TE buffer (see Recipes)
- Elution buffer (see Recipes)
Equipment
- Sonicator (Branson Digital Sonifier 450) (Branson Ultrasonics Corp)
- Centrifuge
- 15 ml conical tubes
- T75 flasks
- Cell scrapers
Procedure
Outline
- Three days before starting this procedure, usually on a Friday, split your cells 1:4 so that they will be confluent on Monday.
- Day 1 (Monday)-split your cells.
- Day 2 (Tuesday)-treat your cells with whatever compound you have found that causes transcriptional activation of your gene of interest. Fix cells when you believe p65 will be at the regulatory region of your gene of interest. Count cells and aliquot 4 million cells per 15 ml conical tube.
- Day 3 (Wednesday)-Sonicate samples, pre-clear samples, and IP with p65 antibody overnight.
- Day 4 (Thursday)-Wash samples, elute, and reverse crosslink.
- Day 5 (Friday)-Do phenol/chloroform extraction and ethanol precipitation. Proceed to do PCR of your region of interest if you have time that day.
Note: First determine the sonication conditions for your particular cell type to yield DNA fragments between 300-1,000 bp. This is usually around 12% output, 5 cycles of 10 sec each with a 30 sec cooling period in between sonications. A 1.2% agarose Ethidium bromide gel of the DNA will show a brighter smear in this region, as shown in the second lane of Figure 1.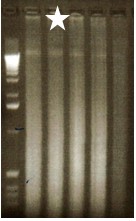
Figure 1. Determining sonication conditions for HCT116 for ChIP. The lane denoted with a star shows the proper sonication conditions.
Day 1
Split your cells. Cells should be in log phase of growth. Usually I use a T75 flask to grow the cells. The cells should be about 60-70% confluent for the next day. If you split a confluent T75 flask of your cells with 3 ml trypsin + 9 ml media, you will need 4 ml cells (~5 x 106 cells) for each T75 (you can do 3 total, so untreated, treated, and a counting dish per T75 flask). These conditions are for HCT116, so adjust accordingly for your cell line.
Day 2
- Treat your cells the next day with DMSO or PBS for control (depending on your compound’s solubility) and the compound of interest. To detect p65 on the PUMA promoter after sorafenib treatment, I did an 8 h time point. This may change depending on the transcription rate of your protein. I would use the time that you first started seeing your protein expression increase.
- While you wait for your time point, count the cells in your extra flask. This will tell you how many cells/ml you have in your other flasks. If you can’t do this due to limited sample amounts, after the time point for your compound, and before you fix the cells, count them.
- Add formaldehyde to a final concentration of 1% in your flask. Let sit at room temperature (RT) with gentle rocking for 15 min.
- Add 1 M glycine to a final concentration of 125 mM to stop the reaction. Mix gently.
- Using a cell scraper, scrape off the cells and spin down cells at 400 x g for 5 min at 4 °C. Aspirate supernatant.
- Resuspend cells in cold PBS. Make aliquots of 4 million cells per 15 ml conical tube. Label tubes correctly. Spin down again and aspirate supernatant. Snap freeze in liquid nitrogen or a dry ice/100% ethanol bath, and store at -80 °C.
Day 3
- Remove 2 untreated and 2 treated tubes from the -80 °C freezer. Place on ice to thaw.
Note: All tubes on ice from now on.
- Resuspend cell pellet in 200 μl cold EBC buffer with protease inhibitors (complete mini EDTA-free tablet, Roche. I make up 10 ml of EBC buffer with 1 tablet of the complete mini EDTA-free protease inhibitor cocktail). One set of untreated and treated will be used for IP for p65 and the other will be a minus antibody (-Ab) control. So you will have 4 tubes total and will need 800 μl cold EBC buffer+protease inhibitors.
- Sonicate samples at 12% output, 10 sec x 5 cycles, with 30 sec on ice between sonications.
- Spin cells down at 9,000 x g for 10 min.
- Remove supernatant into a fresh tube.
- Follow the protocol for the Chromatin Immunoprecipitation Assay Kit.
- Make a master mix of protease inhibitor cocktail solution with ChIP dilution buffer (included in the kit). Since there are 4 total samples, you will need 6.4 ml ChIP dilution buffer and 800 μl of the 10x protease inhibitor cocktail solution. Add 1,800 μl to each sample. Remove 20 μl from the untreated and treated –Ab tubes as input controls and save at 4 °C for later use.
- Preclear the lysates with 75 μl Protein A/G PLUS-agarose for 1 h rotating at 4 °C.
- Pellet at 14,000 x g for ~15 sec. Save the supernatant to a fresh tube, being careful to not disturb the pellet. I usually leave about 50 μl of supernatant behind so as not contaminate the supernatant with the agarose. Keep the tubes with the supernatant, toss the pellets in the trash.
- For the –Ab tubes, rotate at 4 °C overnight. For the p65 IPs, use 2 μg of p65 antibody per tube. Rotate overnight at 4 °C.
- Make a master mix of protease inhibitor cocktail solution with ChIP dilution buffer (included in the kit). Since there are 4 total samples, you will need 6.4 ml ChIP dilution buffer and 800 μl of the 10x protease inhibitor cocktail solution. Add 1,800 μl to each sample. Remove 20 μl from the untreated and treated –Ab tubes as input controls and save at 4 °C for later use.
Day 4
- Add 60 μl of Protein A/G PLUS-agarose to all 4 tubes (–Ab tubes. Rotate for 1 h at 4 °C to pull down immuno complexes.
- Briefly centrifuge pellets at 14,000 x g, 15 sec. Remove supernatant and discard.
- Wash pellet with the following list of buffers that are included in the kit. If you do not have the kit, the recipe is included. Centrifuge after washing, remove supernatant completely and resuspend in the next buffer on this list. The wash buffers increase in salt concentration to remove non-specific binding to the antibody/agarose complex. Use 500 μl of the buffer, rotating for 5 min at 4 °C for each wash.
- 1x low salt immune complex wash buffer.
- 1x high salt immune complex wash buffer.
- 1x LiCl immune complex wash buffer.
- 2x TE buffer.
- 1x low salt immune complex wash buffer.
- Freshly prepare elution buffer. Elute protein by adding 250 μl of elution buffer, vortexing, and incubating at RT for 15 min with rotation. Briefly spin, remove eluate to a new tube, and repeat elution with another 250 μl elution buffer. Combine eluents for a final volume of 500 μl per IP reaction (at this point you should have 4 tubes, 2 for p65 IP, 2 for -Ab, with control and treated for each).
- Add 20 μl 5 M NaCl to the eluents. Also, add 5 μl 5 M NaCl to the input controls and heat all samples at 65 °C for 4 h to reverse the crosslinking. At this step the samples can be stored at –20 °C, or proceed to DNA extraction if you have time.
Day 5
- Recover DNA by phenol/chloroform extraction.
- Measure the volume of the reversed-cross linked samples. Add an equal volume of phenol to each sample, vortex, and spin for 3 min at 14,000 x g. Transfer supernatant to a fresh tube (avoid the bottom and interphase layer).
- Add an equal volume of chloroform, vortex, and spin for 3 min at 14,000 x g. Transfer supernatant to a fresh tube (avoid the bottom and interphase layer).
- Measure the volume of the reversed-cross linked samples. Add an equal volume of phenol to each sample, vortex, and spin for 3 min at 14,000 x g. Transfer supernatant to a fresh tube (avoid the bottom and interphase layer).
- Precipitate DNA by ethanol precipitation.
- Measure the volume of the extracted samples. Add 0.1 volume 3 M sodium acetate, 2 volumes of 100% ethanol, and 4 μl of 5 mg/ml glycogen (as a DNA carrier) to each tube.
- Vortex and place at -20 °C for 30 min.
- Spin for 20 min at 14,000 x g at 4 °C.
- Very carefully pour off the supernatant, watching the DNA pellet very carefully so that it does not dislodge.
- Wash pellets with 500 μl 70% ethanol. Vortex and spin for 5 min at 14,000 x g at 4 °C.
- Very carefully pour off the supernatant, watching the DNA pellet so that it does not dislodge. It will be very “slippery” at this point. I use an unfiltered tip to wick away the remaining ethanol from the pellet and to push the pellet back to the bottom of the tube if it has moved.
- Flip the tube upside-down and air dry for 7 min.
- Resuspend in 30 μl water.
- Measure the volume of the extracted samples. Add 0.1 volume 3 M sodium acetate, 2 volumes of 100% ethanol, and 4 μl of 5 mg/ml glycogen (as a DNA carrier) to each tube.
- Proceed with PCR using primers surrounding the potential binding site of interest in the gene promoter. Product size of DNA should be around 100-200 bp. Run PCR reaction on a 2% TBE or TAE gel and document.
Recipes
- EBC buffer
50 mM Tris (pH 7.5)
100 mM NaCl
0.5% NP-40 - ChIP dilution buffer
0.01% SDS
1.1% Triton X-100
1.2 mM EDTA
16.7 mM Tris-HCl (pH 8.1)
167 mM NaCl - 10x protease inhibitor solution
Dissolve one complete mini EDTA-free tablet in 1 ml water. - Low salt immune complex wash buffer
0.1% SDS
1% Triton X-100
2 mM EDTA
20 mM Tris-HCl (pH 8.1)
150 mM NaCl - High salt immune complex wash buffer
0.1% SDS
1% Triton X-100
2 mM EDTA
20 mM Tris-HCl (pH 8.1)
500 mM NaCl - LiCl immune complex wash buffer
0.25 M LiCl
1% IGEPAL-CA630
1% deoxycholic acid (sodium salt)
1 mM EDTA
10 mM Tris (pH 8.1) - TE buffer
10 mM Tris-HCl
1 mM EDTA (pH 8.0) - Elution buffer
1% SDS
0.1 M NaHCO3
Acknowledgments
This protocol was adapted from Dudgeon et al. (2012). This work was supported by NIH grants CA106348 and CA121105 and American Cancer Society grant RSG-07-156-01-CNE (LZ); Flight Attendant Medical Research Institute, NIH grant CA129829 and American Cancer Society grant RGS-10-124-01-CCE (JY); NIH National Research Service Award postdoctoral fellowship grant F32CA139882 (CD); and China Scholarship Council (RP). LZ is a scholar of the V Foundation for Cancer Research.
References
- Dudgeon, C., Peng, R., Wang, P., Sebastiani, A., Yu, J. and Zhang, L. (2012). Inhibiting oncogenic signaling by sorafenib activates PUMA via GSK3β and NF-κB to suppress tumor cell growth. Oncogene 31(46): 4848-4858.
- Wang, P., Yu, J. and Zhang, L. (2007). The nuclear function of p53 is required for PUMA-mediated apoptosis induced by DNA damage. Proc Natl Acad Sci U S A 104(10): 4054-4059.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Dudgeon, C. R. (2013). p65 Chromatin Immunoprecipitation Protocol. Bio-protocol 3(8): e683. DOI: 10.21769/BioProtoc.683.
Category
Molecular Biology > DNA > DNA-protein interaction
Biochemistry > Protein > Immunodetection > ChIP
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









