- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Trimolecular Fluorescence Complementation (TriFC) Assay for Direct Visualization of RNA-Protein Interaction in planta
Published: Vol 7, Iss 20, Oct 20, 2017 DOI: 10.21769/BioProtoc.2579 Views: 12899
Reviewed by: Tie LiuAlexandros AlexandratosHonghong Wu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Analysis of RNA-protein Interactions Using Electrophoretic Mobility Shift Assay (Gel Shift Assay)
Saiprasad Goud Palusa and Anireddy S. N. Reddy
Nov 20, 2013 31366 Views

Plant ARGONAUTE Protein Immunopurification for Pathogen Cross Kingdom Small RNA Analysis
Florian Dunker [...] Arne Weiberg
Feb 5, 2021 6375 Views

Turbo-RIP: A Protocol for TurboID-based RNA Immunopurification to Map RNA Landscapes in Plant Biomolecular Condensates
Zhi Zhang [...] Panagiotis Nikolaou Moschou
Feb 5, 2026 330 Views
Abstract
RNA-Protein interactions play important roles in various eukaryotic biological processes. Molecular imaging of subcellular localization of RNA/protein complexes in plants is critical for understanding these interactions. However, methods to image RNA-Protein interactions in living plants have not yet been developed until now. Recently, we have developed a trimolecular fluorescence complementation (TriFC) system for in vivo visualization of RNA-Protein interaction by transient expression in tobacco leaves. In this method, we combined conventional bimolecular fluorescence complementation (BiFC) system with MS2 system (phage MS2 coat protein [MCP] and its binding RNA sequence [MS2 sequence]) (Schonberger et al., 2012). Target RNA is tagged with 6xMS2 and MCP and RNA binding protein are fused with YFP fragments. DNA constructs encoding such fusion RNA and proteins are infiltrated into tobacco leaves with Agrobacterium suspensions. RNA-Protein interaction in vivo is observed by confocal microscope.
Keywords: Long non-coding RNABackground
Recently, a variety of types of long-noncoding RNAs (lncRNAs) has been identified and shown to play important roles in transcriptional regulation and chromatin modification (St Laurent et al., 2015). So far, most of the molecular mechanisms for lncRNA-mediated functions are closely related with RNA-Protein interactions (St Laurent et al., 2015). Therefore, an experiment for RNA-Protein interaction is a key step in functional study of lncRNAs. In plants, molecular functions of lncRNAs are only beginning to be characterized, and the molecular basis of lncRNA-mediated gene regulation is still poorly understood. Though techniques for RNA visualization in plants have been well developed, visual assay for RNA-Protein interaction in plant is still poorly developed (Christensen et al., 2010). To develop the visual assay for RNA-Protein interaction in plants, we modified and combined MS2 system for RNA imaging technique with conventional BiFC system for protein-protein interaction (Schonberger et al., 2012) (Figure 1D). We generated binary Gateway vectors (pBA3130, 3132, 3134, and 3136) for transient BiFC assay (Seo et al., 2017) and got binary Gateway vectors (pBA-GW-6xMS2 and pBA-6xMS2-GW) for RNA tagging from Dr. Ulrich Z. Hammes (Schonberger et al., 2012). We tested and confirmed this TriFC assay was working well in plants with lncRNA, ELENA1, and MED19a protein (Seo et al., 2017). TriFC assay in plants will provide new insights in interaction between lncRNAs and proteins.
Materials and Reagents
- Pipette tips (Thermo Fisher Scientific, Fisher ScientificTM BasixTM Universal Tips)
- Fisherbrand sterile 100 x 15 mm polystyrene Petri dish (Fisher Scientific, catalog number: FB0875713 )
- 1 ml syringes (BD, catalog number: 302100 )
- 50 ml Falcon tubes (Corning, Falcon®, catalog number: 352070 )
- Agrobacterium tumefaciens (strain GV3101)
- Nicotiana benthamiana (N. benthamiana) plants; 2-4 weeks old (6-10 leaves stage)
- Gateway entry clones for RNA, RNA binding protein (RNA-BP), and MCP
- TriFC Gateway destination vectors (pBA3130, 3132, 3134, 3136, pBA-GW-MS2, and pBA-MS2-GW)
- LR clonase II (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11791100 )
- LB medium powder (MP Biomedicals, catalog number: 113002082 )
- Spectinomycin (1,000x; 100 mg/ml) (Sigma-Aldrich, catalog number: S4014 )
- Gentamycin (1,000x; 50 mg/ml) (Sigma-Aldrich, catalog number: G1264 )
- Kanamycin (1,000x; 100 mg/ml) (Sigma-Aldrich, catalog number: K0200000 )
- Bacto agar (BD, BactoTM, catalog number: 214010 )
- Ethanol or DMSO
- MES (Sigma-Aldrich, catalog number: M8250 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- Acetosyringone (Sigma-Aldrich, catalog number: D134406 )
- LB media (see Recipes)
- LB agar media (see Recipes)
- 100 mM acetosyringone stock (see Recipes)
- LB-MES (pH 5.6) (see Recipes)
- Resuspension solution (see Recipes)
Equipment
- PIPETMAN ClassicTM Pipettes (Gilson, model: P1000, P100, P20, catalog number: F123602 , F123615 , F123600 )
- Centrifuge for 50 ml tubes (Beckman Coulter, model: Avanti® J-20XP )
- Spectrometer (Biochrom, model: Ultrospec 2100 pro )
- Confocal laser scanning microscope (ZEISS, model: LSM 780 )
- Autoclave (TOMY DIGITAL BIOLOGY, model: ES-215 )
- Laminar flow cabinet (NuAire, model: NU-440-400E )
- Incubator (MMM Medcenter Einrichtungen, model: INCUCELL 55 )
- Shaking incubator (Infors, model: Multitron Standard )
Software
- ZEN (Image analysis program for ZEISS confocal microscope)
Procedure
- Construct TriFC vectors by LR reactions (Gateway) with your entry clones (RNA, RNA-BP, and MCP) following the manufacturer’s instruction; LR reaction MCP and RNA-BP entry clones with Gateway BiFC vectors (pBA3130, 3132, 3134, and 3136; Figures 1A and 1B) (Seo et al., 2017). LR reaction RNA entry clones with Gateway MS2 tagging vectors (pBA-6xMS2-GW or pBA-GW-6xMS2; Figure 1C) (Schonberger et al., 2012). In this study, we used pENTR-ELENA1 for RNA entry clone, pENTR-MED19a for RNA-BP entry clone, and pENTR-MCP for MCP entry clone.
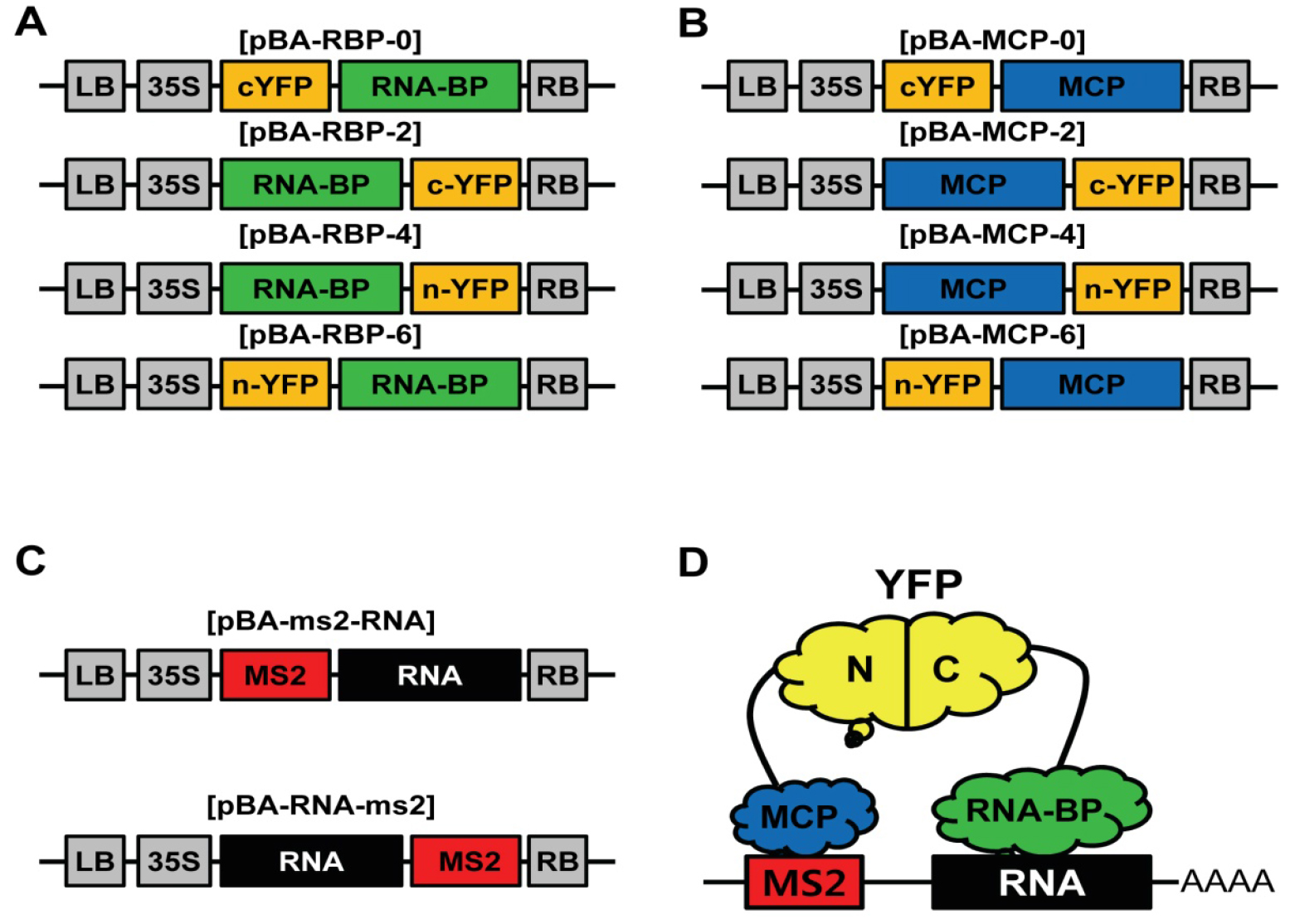
Figure 1. Schematic representation of the vectors for TriFC system. A. Schematic diagram of expression vectors of RNA binding protein fused with YFP fragments; B. Schematic diagram of expression vectors of MCP fused with YFP fragments; C. Schematic diagram of MS2 tagged RNA expression vectors; D. Illustration of the TriFC system for RNA-Protein interaction.
- Transform 50 µl Agrobacterium competent cells (1010 cfu/ml) with each DNA construct (50 ng) (3 different vectors; Figures 1A, 1B, and 1C) by heat shock method (Höfgen and Willmitzer, 1988).
- Spread the cells on the LB/spectinomycin agar media (see Recipes) and grow for 2 days in an incubator at 28-30 °C.
- Inoculate one single colony of Agrobacterium in 5 ml LB with appropriate antibiotics (see Recipes). Grow overnight in a shaking incubator with 200 rpm at 28-30 °C.
- Use 250 μl of the overnight culture to inoculate 10 ml LB-MES (pH 5.6) (see Recipes) (with same antibiotics) plus 4 μl of 100 mM acetosyringone (see Recipes) and grow overnight in the shaking incubator with 200 rpm at 28-30 °C.
- Measure the A600 of overnight cultures (OD = 0.8-1.0).
- Collect bacterial cells (4,000 x g, 10 min) at room temperature and resuspend the pellet in resuspension solution (see Recipes). Adjust the final A600 to 0.4.
- Leave on a bench (room temperature) for over 4 h (or overnight) before infiltration.
- Transfer 1 ml of each Agrobacterium cell suspension into a 10 ml tube and mix well (Figure 2A).
- Perform the infiltration with a 1 ml syringe. Simple press the syringe (no needle) on the underside of a leaf (Note: Avoid cotyledons.), and exert a counter-pressure with finger on the other side. Successful infiltration is often observed as a spreading of ‘wetting’ area in the leaf and label the infiltrated region. (Figure 2B).
- Observe the YFP signals under a confocal laser scanning microscope 2-3 days after infiltration (Figure 2C). Excitation wavelength is 514 nm and detection rage of emission wavelength is 520-550 nm.

Figure 2. Transient expression for TriFC assay in N. benthamiana. A. Mix three different Agrobacterium suspensions. 1 carries the RNA-BP construct (Figure 1A), 2 carries the MCP construct (Figure 1B), and 3 carries the MS2 tagged RNA construct (Figure 1C). B. Infiltration in tobacco leaves with a syringe; C. Observe the infiltrated region with a confocal microscope 2-3 days after infiltration.
Data analysis
- To avoid false positive signal, we checked MED19a and MCP interaction (Figure 3 [top]), and MED19a, MCP, and antisense ELENA1 combination (Figure 3 [middle]) as negative controls. We confirmed that there was no YFP signal in those negative controls (we tested three independent plants).
- We could observe YFP signal in MED19a, MCP, and sense ELENA1 combination (Figure 3 [bottom]). We tested three independent plants and observed YFP signals in all the plants.
- Detailed information about ELENA1 and MED19a is described in (Seo et al., 2017).
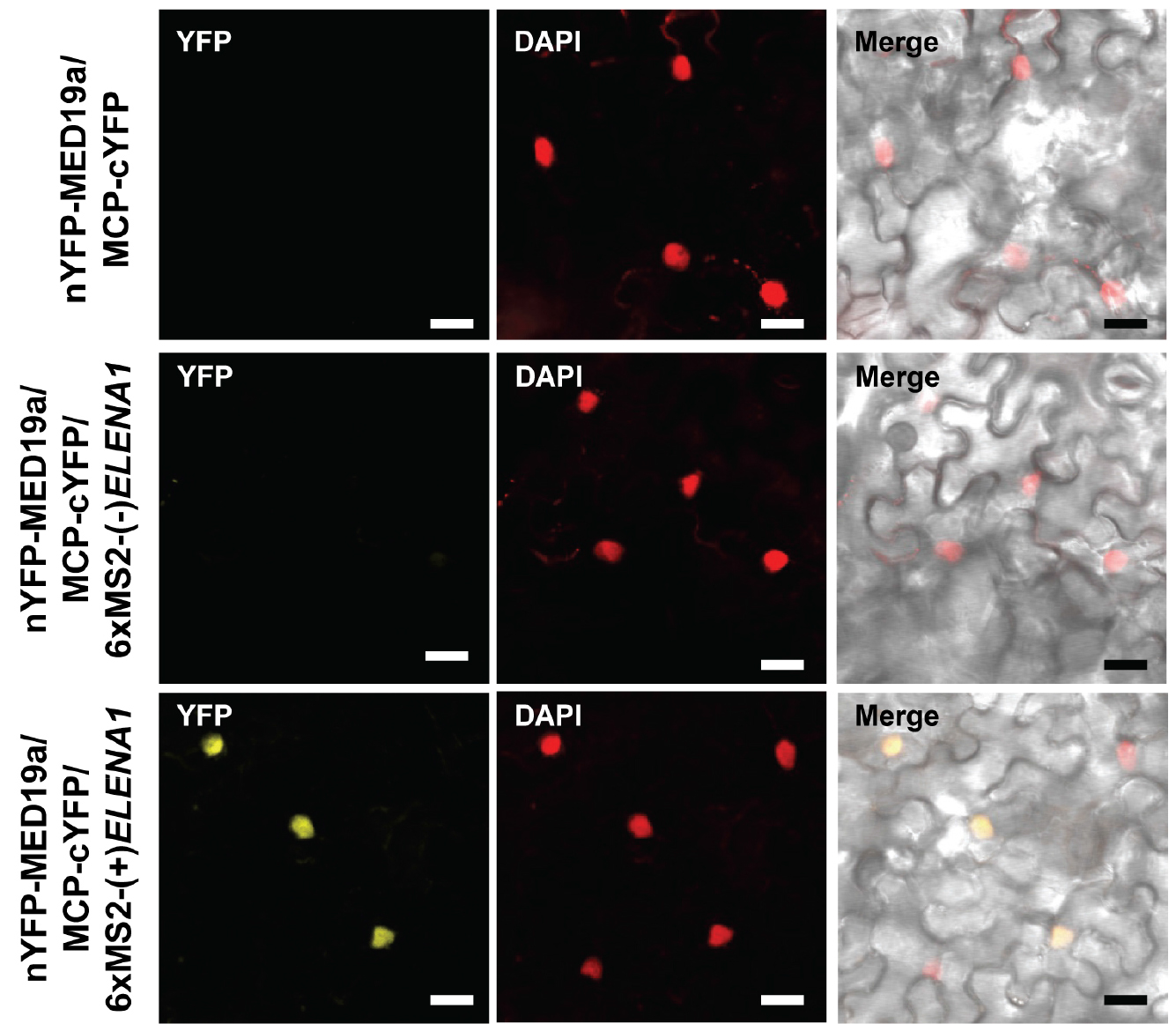
Figure 3. Confocal microscopy images of TriFC assay. nYFP was fused to MED19a and cYFP was fused to MCP. 6xMS2 nucleotide sequences fused to sense or anti-sense ELENA1. Confocal images were taken 3 days after infiltration. Scale bars = 20 μm.
Notes
- An optimal combination of 6xMS2 tag, nYFP, and cYFP orientation is sometimes critical for the TriFC assay. Therefore, other combinations should be tried if the signal is not detected.
- Post-transcriptional gene silencing (PTGS) is a major cause for the lack of efficiency in transient expression experiments (Voinnet et al., 2003). Therefore, we usually co-infiltrate UBQ:p19, which suppresses PTGS, together with other constructs as it greatly improves the efficiency.
Recipes
- LB media
LB media with appropriate antibiotics (kanamycin [50 µg/ml] for MS2 tagging vector [Figure 1C] and spectinomycin [50 µg/ml] for partial YFP fused vectors [Figures 1A and 1B])
- LB agar media
LB media with 2% Bacto agar and appropriate antibiotics (kanamycin [50 µg/ml] for MS2 tagging vector [Figure 1C] and spectinomycin [50 µg/ml] for partial YFP fused vectors [Figures 1A and 1B])
- 100 mM acetosyringone stock
100 mM acetosyringone stock in ethanol or DMSO, stored at -20 °C
- LB-MES (pH 5.6)
LB with 10 mM MES (pH 5.6)
- Resuspension solution
10 mM MgCl2
10 mM MES-KOH (pH 5.6)
100 μM acetosyringone
Note: Added after autoclaving and immediately before using.
Acknowledgments
We thank Dr. Ulrich Z. Hammes for the Gateway 6xMS2 tagging vectors. This work was supported by Singapore NRF RSSS Grant (NRF-RSSS-002).
References
- Christensen, N. M., Oparka, K. J. and Tilsner, J. (2010). Advances in imaging RNA in plants. Trends Plant Sci 15(4): 196-203.
- Höfgen, R. and Willmitzer, L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16(20): 9877.
- Schonberger, J., Hammes, U. Z. and Dresselhaus, T. (2012). In vivo visualization of RNA in plants cells using the λN22 system and a GATEWAY-compatible vector series for candidate RNAs. Plant J 71(1): 173-181.
- Seo, J. S., Sun, H. X., Park, B. S., Huang, C. H., Yeh, S. D., Jung, C. and Chua, N. H. (2017). ELF18-INDUCED LONG-NONCODING RNA associates with mediator to enhance expression of innate immune response genes in Arabidopsis. Plant Cell 29(5): 1024-1038.
- St Laurent, G., Wahlestedt, C. and Kapranov, P. (2015). The Landscape of long noncoding RNA classification. Trends Genet 31(5): 239-251
- Voinnet, O., Rivas, S., Mestre, P. and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33(5): 949-956.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Seo, J. S. and Chua, N. (2017). Trimolecular Fluorescence Complementation (TriFC) Assay for Direct Visualization of RNA-Protein Interaction in planta. Bio-protocol 7(20): e2579. DOI: 10.21769/BioProtoc.2579.
Category
Plant Science > Plant molecular biology > RNA > RNA-protein interaction
Molecular Biology > RNA > RNA-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










