- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Differentiation of Myeloid-derived Suppressor Cells from Murine Bone Marrow and Their Co-culture with Splenic Dendritic Cells
Published: Vol 7, Iss 18, Sep 20, 2017 DOI: 10.21769/BioProtoc.2558 Views: 12329
Reviewed by: Ivan ZanoniMeenal SinhaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Differentiation, Maintenance, and Contraction Profiling of Human Induced Pluripotent Stem Cell–Derived Cardiomyocytes
Matthijs Snelders [...] Jeroen Essers
Mar 5, 2025 3937 Views

Isolation and Culture of Ferret Airway Stem Cells
Ziying Yan [...] Feng Yuan
Jul 20, 2025 2413 Views

Optimization of Adipogenic Differentiation Protocol for Murine and Human Cell Culture Models
Junwan Fan [...] Wenyan He
Jan 20, 2026 230 Views
Abstract
Myeloid-derived suppressor cells (MDSCs) possess the ability to suppress the immune response, and to amplify the regulatory properties of other immune cells, i.e., dendritic cells. Here we describe a protocol in which MDSCs were differentiated from murine bone marrow cells, and CD11c+ dendritic cells were purified from murine spleens. MDSCs and CD11c dendritic cells can be co-cultured and the immunoregulatory phenotype of the MDSCs-conditioned dendritic cells could be assessed by means of a specific functional in vivo experiment, i.e., a skin test as a measure of the delayed-type hypersensitivity reaction toward a poorly immunogenic antigen.
Keywords: Myeloid-derived suppressor cellsBackground
The myeloid-derived suppressor cells (MDSCs) are a group of myeloid cells comprised of precursor of macrophages, granulocytes, dendritic cells and myeloid cells at earlier stages of differentiation (Youn et al., 2008) accumulating in large numbers in lymphoid tissues of tumor-bearing mice as well as in mice with infectious diseases, sepsis and trauma. The main feature of these cells is their ability to suppress T cell responses in Ag-specific and/or nonspecific fashion. These cells are now considered as one of the major cell type responsible for tumor-associated immune defects; main factors implicated in MDSC-mediated immune suppression include high expression of Arg1 (Marvel and Gabrilovich, 2015). Arginase 1 (Arg1) and indoleamine 2,3-dioxygenase 1 (IDO1) are immunoregulatory enzymes catalyzing the degradation of L-arginine (L-Arg) and L-tryptophan (L-Trp), respectively, resulting in local amino acid deprivation. In addition, unlike Arg1, IDO1 is also endowed with non-enzymatic signaling activity in dendritic cells (DCs) (Mondanelli et al., 2017). In addition to their inherent immunosuppressive activity, MDSCs might amplify regulatory properties of other immune cells, particularly in tumor microenvironments. Although some mechanisms underlying MDSC-macrophage interaction have been established (Ugel et al., 2015), the cross-talk between MDSCs and DCs is still unclear (Ostrand-Rosenberg et al., 2012); to fill this gap, we have developed this protocol and we demonstrated that Arg1+ MDSCs confer to DCs an IDO1-dependent, immunosuppressive phenotype via Arg1 metabolites (i.e., polyamines such as putrescine and spermidine) (Mondanelli et al., 2017). The Arg and Trp immunoregulatory pathways are functionally integrated, this integration occurring both intra- (i.e., DCs) and inter-cellularly (MDSCs and DCs) (Mondanelli et al., 2017).
Materials and Reagents
- Petri dishes (Corning, Falcon®, catalog number: 351029 )
- 15 ml Falcon tubes (Corning, Falcon®, catalog number: 352096 )
- Cell strainers (Corning, Falcon®, catalog number: 352340 )
- 5 ml sterile pipettes (Corning, Falcon®, catalog number: 357543 )
- 10 ml sterile pipettes (Corning, Falcon®, catalog number: 357551 )
- 200 µl sterile tips (Biotix, Neptune®, catalog number: 2102 NS )
- 1,000 µl sterile tips (Biotix, Neptune®, catalog number: 2372 S )
- 24-well plates (Corning, Falcon®, catalog number: 353047 )
- 50 ml Falcon tubes (Corning, Falcon®, catalog number: 352070 )
- 10 ml syringe (Terumo Medical, catalog number: SS+10S21381 )
- 1 ml syringe with 26 G ½ needle (Terumo Medical, catalog number: SS+01H26131 )
- 2 ml syringe plunger (Terumo Medical, catalog number: SS-02S2238 )
- 6-well plates (Corning, Falcon®, catalog number: 353046 )
- Permeable support for 24-well plate with 0.4 μm translucent high density PET membrane (Corning, Falcon®, catalog number: 353495 )
- LS columns (Miltenyi Biotec, catalog number: 130-042-401 )
- MS columns (Miltenyi Biotec, catalog number: 130-042-201 )
- Pasteur pipette (Sigma-Aldrich, catalog number: Z627992-1000EA )
- C57BL/6 female mice, 6 weeks old (C57BL/6NCrl) (Charles River Laboratories, catalog number: 027 )
- RPMI 1640 medium (Thermo Fisher Scientific, catalog number: 11875093 )
- FCS (Thermo Fisher Scientific, catalog number: A3160801 )
- L-Glutamine (Thermo Fisher Scientific, catalog number: 25030024 )
- Penicillin-streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- HEPES (Thermo Fisher Scientific, catalog number: 15630056 )
- 2-Mercaptoethanol (Thermo Fisher Scientific, GibcoTM, catalog number: 31350010 )
- Trypan blue (Thermo Fisher Scientific, GibcoTM, catalog number: 15250061 )
- Phosphate buffered saline (PBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10010023 )
- Bovine serum albumin (BSA) (Rockland Immunochemicals, catalog number: BSA-50 )
- Ethylenediaminetetraacetate acid (EDTA) (AppliChem, catalog number: 131669.1211 )
- CD11b MicroBeads, human and mouse (Miltenyi Biotec, catalog number: 130-049-601 )
- CD11c MicroBeads UltraPure, mouse (Miltenyi Biotec, catalog number: 130-108-338 )
- HBSS, no calcium, no magnesium (Thermo Fisher Scientific, catalog number: 14170088 )
- Sodium chloride (NaCl) (CARLO ERBA Reagents, catalog number: 479687 )
- Tris (Bio-Rad Laboratories, catalog number: 1610719 )
- Potassium chloride (KCl) (CARLO ERBA Reagents, catalog number: 471177 )
- Recombinant murine GMCSF (PeproTech, catalog number: 315-03 )
- Recombinant murine IL-4 (PeproTech, catalog number: 214-14 )
- Collagenase from Clostridium histolyticum (Sigma-Aldrich, catalog number: C5138-1G )
- Histodenz (Sigma-Aldrich, catalog number: D2158-100G )
- Nor-NOHA (Cayman Chemicals, catalog number: 10006861 )
- MACS buffer (see Recipes)
- RPMI medium (see Recipes)
- TCCM (see Recipes)
- Collagenase 100 U/ml and 400 U/ml (see Recipes)
- Nycodenz (see Recipes)
- Nycodenz buffer (see Recipes)
Equipment
- Scissor (Isolab Laborgeräte, catalog number: 048.25.130 )
- Sterile biosafety cabinet
- P20L pipette (Pipetman L) (Gilson, catalog number: FA10003M )
- P200L pipette (Pipetman L) (Gilson, catalog number: FA10005M )
- P1000L pipette (Pipetman L) (Gilson, catalog number: FA10006M )
- Pipet controller (Corning, Falcon®, catalog number: 357471 )
- Centrifuge (Eppendorf, model: 5810 R )
- Incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: BB15 )
- -80 °C freeze (Thermo Fisher Scientific, Thermo ScientificTM, model: FormaTM 88000 Series )
- Light microscope (ZEISS Primostar)
- MiniMACS separator (Miltenyi Biotec, catalog number: 130-042-102 )
- MidiMACS separator (Miltenyi Biotec, catalog number: 130-042-302 )
Procedure
- MDSCs isolation from bone marrow
- Take the hind limbs from two C57BL/6 mice, cutting the bone extremities.
- Under a sterile biosafety cabinet, completely remove flesh from the bones. Put the bones in a Petri dish and inject 10 ml of complete RPMI medium (see Recipes) inside each thigh bone using a 10 ml syringe with a 26 G ½ needle, in order to extract the bone marrow.
- Aspirate and discharge the medium until the bone marrow cells are completely resuspended, using the same initial 10 ml of RPMI.
- Transfer the bone marrow cells suspension in a 15 ml Falcon tube supplied with a 40 µm cell strainer, then remove the cell strainer and centrifuge (582 x g, 10 min, RT).
- Discard the supernatant and resuspend cells in 10 ml of fresh complete RPMI.
- Count cells, properly diluting them in trypan blue (from 4 legs, approximately 50 x 106 cells can be obtained).
- Centrifuge the cell suspension (582 x g, 10 min, RT).
- Sort CD11b+ cells:
- Resuspend cell pellet in 80 µl of ice-cold MACS buffer (see Recipes) per 107 total cells.
- Add 20 µl of CD11b Microbeads per 107 total cells.
- Mix well and incubate for 15 min at 4 °C.
- Wash cells by adding 1-2 ml of ice-cold MACS buffer per 107 cells and centrifuge (582 x g, 10 min, 4 °C).
- Resuspend up to 108 cells in 500 µl of MACS buffer. Proceed with magnetic separation.
- Place the column in the magnetic field and rinse with the appropriate amount of buffer: MS column 500 µl (up to 108 cells), LS column 3 ml (from 108 cells to 109 cells).
- Apply the cell suspension onto the column. Collect unlabeled cells which pass through and wash column with the appropriate amount of buffer. Perform washing steps by adding ice-cold MACS buffer three times, each time once the column reservoir is empty (MS column: 3 x 500 µl, LS column: 3 x 3 ml). Collect the total effluent (the unlabeled cell fraction).
- Remove column from the separator and place it on a suitable collection tube.
- Pipette appropriate amount of buffer onto the column. Immediately flush out fraction with the magnetically labeled cells by firmly applying the plunger supplied with the column (MS: 1 ml, LS: 5 ml).
- Centrifuge the labeled cells (CD11b+ cells) (582 x g, 10 min, RT). Usually, from 50 x 106 BM cells, the recovery of CD11b+ cells is about 15 x 106 cells.
- Resuspend cell pellet in 80 µl of ice-cold MACS buffer (see Recipes) per 107 total cells.
- Resuspend the sorted CD11b+ cells (1 x 106 cells/ml) in complete RPMI containing 30% TCCM (see Recipes), 20 ng/ml of GMCSF and 20 ng/ml of IL-4. Both GMCSF and IL-4 are resuspended in RPMI and aliquoted and stored at -80 °C.
- Plate the cells 1 ml/well in a 24-well plate. Incubate cells for 4 days at 37 °C.
- After 4 days change the medium: recover cells by pipetting them in a 50 ml Falcon tube and centrifuge (805 x g, 10 min, RT). Discard the supernatant and add new RPMI containing 30% TCCM, 20 ng/ml of GMCSF and 20 ng/ml of IL-4. Plate again the cells 1 ml/well in a 24-well plate.
- After 7 days, isolate CD11c+ DCs from spleens.
- CD11c+ DCs isolation from spleen
- Before starting, prepare 2 solutions of collagenase; for each spleen prepare 1 ml of each collagenase solution (15 ml of 100 and 15 ml of 400 U/ml) (see Recipes) as described in the Recipes section.
- Take spleens from 15 C57BL/6 mice.
- Put the spleens in a Petri dish and inject in each spleen 1 ml of the 100 U/ml collagenase solution (i.e., for 15 spleens, use 15 ml of 100 U/ml collagenase solution) using a 10 ml syringe with a 26 G ½ needle, until the spleen change colour (spleens colour should fade from red to white). Repeat this step twice.
- Recover the cell suspension and filter it in a 50 ml Falcon tube supplied with a 40 µm cell strainer, already containing 5 ml of complete RPMI. Keep this supernatant at RT.
- Incubate the spleens with 1 ml/spleen of 400 U/ml collagenase solution for 30 min at 37 °C.
- Recover the collagenase solution in a 10 ml syringe and homogenize the spleens using a 2 ml syringe plunger.
- Resuspend the spleens completely until all splenic cells are in solution using the collagenase solution recovered before. Be sure that cells are homogeneously resuspended, then filter the cell suspension through the cell strainer onto the same 50 ml Falcon tube already containing the first cell suspension recovered from spleens treated with 100 U/ml collagenase.
- Remove the cell strainer and centrifuge (582 x g, 10 min, RT).
- Discard the supernatant and resuspend the cellular pellet in 15 ml (1 ml/spleen) of a solution composed by 54% of Nycodenz (see Recipes) and 46% of Nycodenz buffer (see Recipes). Divide the obtained cellular suspension in three 15 ml Falcon tubes (5 ml/tube).
- Carefully overlay 3 ml of RPMI medium in each tube containing 5 ml of the cell suspension (Figure 1A).
- Centrifuge at 1,811 x g, 15 min, 4 °C without brake.
- Recover the ring at the interface of the two phases using a Pasteur pipette and transfer the cells into a new 50 ml Falcon tube (Figure 1B).
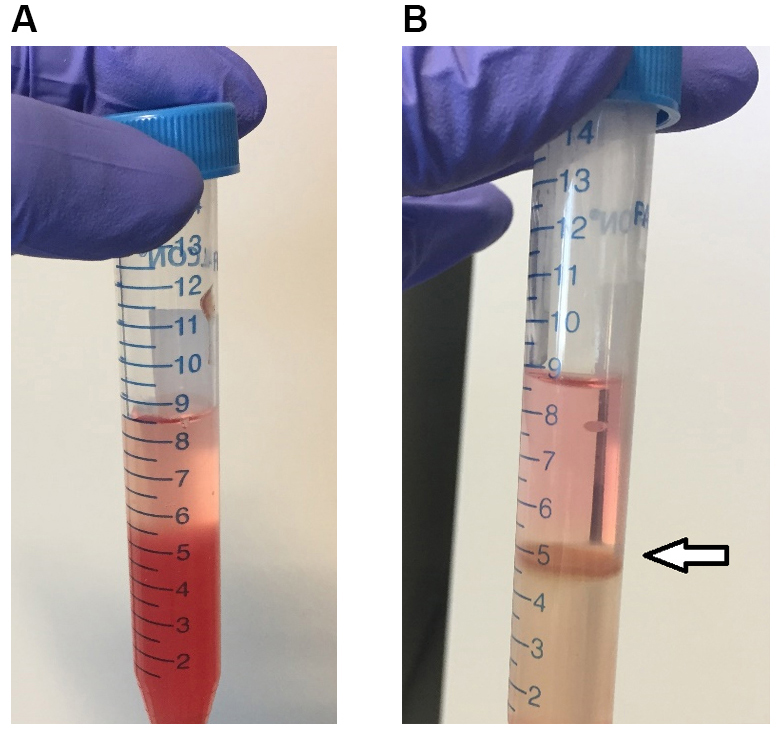
Figure 1. Isolation of dendritic cells from splenocytes by density gradient. A. Density gradient of splenic cells resuspended in Nycodenz (lower phase) and medium RPMI (upper phase) before the centrifugation. B. Density gradient of splenic cells after the centrifugation: the ring at the interface contains CD11c+ dendritic cells. - Add 10 ml of RPMI medium and centrifuge at 582 x g, 10 min, RT. Discard the supernatant and resuspend the cellular pellet in 10 ml of complete RPMI.
- Count cells and centrifuge again at 582 x g, 10 min, RT. Usually, the recovery is about 100 x 106 cells from one spleen.
- Resuspend cell pellet in 8.5 µl of ice-cold MACS buffer per 107 total cells.
- Add 1.5 µl of CD11c Microbeads per 107 total cells.
- Mix well and incubate for 15 min at 4 °C.
- Wash cells by adding 1-2 ml of ice-cold MACS buffer per 107 cells and centrifuge at 582 x g, 10 min, RT.
- Resuspend up to 108 cells in 500 µl of ice-cold MACS buffer. Proceed with magnetic separation.
- Place column in the magnetic field and rinse with the appropriate amount of buffer: MS column 500 µl (up to 108 cells), LS column 3 ml (from 108 cells to 109 cells).
- Apply cell suspension onto the column. Discard unlabeled cells which pass through and wash column with the appropriate amount of buffer. Perform washing steps by adding ice-cold MACS buffer three times, each time once the column reservoir is empty (MS column: 3 x 500 µl, LS column: 3 x 3 ml). Collect the total effluent (the unlabeled cell fraction).
- Remove column from the separator and place it on a suitable collection tube.
- Pipette appropriate amount of buffer onto the column. Immediately flush out fraction with the magnetically labeled cells by firmly applying the plunger supplied with the column (MS: 1 ml, LS: 5 ml).
- Centrifuge the labeled cells (CD11c+ cells) (582 x g, 10 min, RT), discard the supernatant, resuspend cells in 10 ml of complete RPMI and count them. Usually, the recovery of CD11c+ cells is about 1 x 106 cells from one spleen.
- Resuspend cells (1.5 x 106 cells/ml) in complete RPMI and plate 4 ml/well in a 6-well plate. Incubate CD11c+ cells overnight at 37 °C, 5% CO2.
- Co-culture of MDSCs and CD11c+ DCs
For a co-culture of MDSCs and CD11c+ DCs at a 1:2 ratio: - Recover and count CD11c+ cells. Resuspend 1 x 106 cells in 700 µl of complete RPMI. Plate cells in a 24-well plate. Consider 1 x 106 CD11c+ cells for each sample of the co-culture (i.e., CD11c+ alone, CD11c+ co-cultured with untreated MDSCs and CD11c+ co-cultured with MDSCs treated with Nor-NOHA to inhibit Arg1 enzymatic activity).
- Put onto each well containing CD11c+ cells a transwell filled with 100 µl of complete RPMI (without MDSCs) and incubate for 1 h at 37 °C.
- In the meanwhile, incubate MDSCs 1 x 106 cells/ml with the appropriate stimulus (i.e., with and without 150 µM Nor-NOHA to inhibit Arg1 enzymatic activity) for 1 h at 37 °C, then recover and centrifuge MDSCs (582 x g, 10 min, RT).
- Resuspend MDSCs 0.5 x 106 cells in 100 µl of complete RPMI.
- Add 100 µl of MDSCs suspension into the appropriated transwell and incubate overnight at 37 °C.
- Recover MDSCs and CD11c+ cells and supernatants and perform the desired analysis (i.e., a delayed-type hypersensitivity assay using a skin test (Mondanelli et al., 2017) to assess the functional phenotype of CD11c DCs after the co-culture with MDSCs).
Data analysis
Usually, co-culture of MDSCs with DCs must be repeated at least three times to obtain statistically relevant data. Unpaired Student’s t-test was used for in vitro analyses, using at least three values from 2-3 experiments per group (Mondanelli et al., 2017).
Notes
- All steps should be performed under sterile conditions. Even for the isolation of legs and spleens from euthanized mice, use of autoclaved dissection tools and spray ethanol is highly recommended.
- All media should be warmed at 37 °C before use.
- Usually, purity of MDSC is about 60% and purity of CD11c+ cells is about 95%. More in detail, more than 60% of the MDSCs after the TCCM and cytokines stimulation were CD11b and Gr1 positive cells. CD11c+ cells were 90-95% CD11c+, > 95% MHC I-A+, > 95% B7-2+, < 0.1% CD3+, and appeared to consist of 90-95% CD8-, 5-10% CD8+, and 1-5% B220+ PDCA+ (i.e., plasmacytoid DCs or pDCs) cells.
- As reported in literature, there are several methods of MDSCs differentiation, including the use of GCSF, GMCSF and IL-13, or GMCSF and IL-6, or by means of PGE2. We used TCCM, GMCSF and IL-4 because in these specific MDSCs we assessed the highest expression and functional activity of Arg1.
- BMDCs can also be used in this protocol; in order to use the same DCs we used to determine Arg1 expression and function (Mondanelli et al., 2017) we used splenic DCs for the co-culture with MDSCs.
Recipes
Note: All buffers and media should be sterile.
- RPMI medium
Add to RPMI 1640 medium 10% FCS (Thermo Fisher Scientific)
2 mM L-glutamine (stock solution 200 mM)
100 U/ml penicillin-streptomycin (stock solution10,000 U/ml)
10 mM HEPES (stock solution 1 M)
50 μM 2-mercaptoethanol (stock solution 50 mM) - MACS buffer (stored at 4 °C)
Phosphate buffered saline (PBS) pH 7.2
0.5% bovine serum albumin (BSA)
2 mM EDTA - TCCM
- To generate tumor cell conditioned medium (TCMM), subconfluent B16 melanoma cells were kept in RPMI 1640 medium with a reduced (3%) serum concentration for 48 h
- After that time, supernatants were collected, aliquoted and kept at -80 °C until further use (Youn et al., 2008)
- To generate tumor cell conditioned medium (TCMM), subconfluent B16 melanoma cells were kept in RPMI 1640 medium with a reduced (3%) serum concentration for 48 h
- Collagenase 100 U/ml and 400 U/ml
- Prepare a stock solution of collagenase 8,000 U/ml in HBSS w/o Ca and Mg and keep this solution aliquoted (1 ml/tube) at -20 °C
- Starting from the stock solution, dilute collagenase to 100 U/ml in HBSS or 400 U/ml in HBSS and keep these solutions at 4 °C
- Prepare a stock solution of collagenase 8,000 U/ml in HBSS w/o Ca and Mg and keep this solution aliquoted (1 ml/tube) at -20 °C
- Nycodenz (stored at 4 °C)
30.55 g histodenz into 100 ml Milli-Q water (final volume)
Filter through a 0.22 µm filter and protect from light - Nycodenz buffer (aliquoted and stored at -20 °C)
9 g NaCl
0.6055 g Tris
0.22368 g KCl
0.11167 g EDTA
Dissolve in 900 ml Milli-Q water
Adjust pH to 7.5
Bring volume to 1 L
Filter through a 0.22 µm filter, aliquot and store
Acknowledgments
This work was supported by the European Research Council (338954-DIDO; to Prof. Ursula Grohmann and Antonio Macchiarulo) and by Ministero dell’Istruzione, Università e Ricerca, Italy (FIRB RBAP11T3WB; to Prof. Ursula Grohmann, Vincenzo Bronte, and Silvio Bicciato; PRIN 2015C2PP7 to Claudia Volpi). This protocol was adapted from Mondanelli et al. (2017).
References
- Marvel, D. and Gabrilovich, D. I. (2015). Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J Clin Invest 125(9): 3356-3364.
- Mondanelli, G., Bianchi, R., Pallotta, M. T., Orabona, C., Albini, E., Iacono, A., Belladonna, M. L., Vacca, C., Fallarino, F., Macchiarulo, A., Ugel, S., Bronte, V., Gevi, F., Zolla, L., Verhaar, A., Peppelenbosch, M., Mazza, E. M., Bicciato, S., Laouar, Y., Santambrogio, L., Puccetti, P., Volpi, C. and Grohmann, U. (2017). A relay pathway between arginine and tryptophan metabolism confers immunosuppressive properties on dendritic cells. Immunity 46(2): 233-244.
- Ostrand-Rosenberg, S., Sinha, P., Beury, D. W. and Clements, V. K. (2012). Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. Semin Cancer Biol 22(4): 275-281.
- Ugel, S., De Sanctis, F., Mandruzzato, S. and Bronte, V. (2015). Tumor-induced myeloid deviation: when myeloid-derived suppressor cells meet tumor-associated macrophages. J Clin Invest 125(9): 3365-3376.
- Youn, J. I., Nagaraj, S., Collazo, M. and Gabrilovich, D. I. (2008). Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol 181(8): 5791-5802.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mondanelli, G. and Volpi, C. (2017). Differentiation of Myeloid-derived Suppressor Cells from Murine Bone Marrow and Their Co-culture with Splenic Dendritic Cells. Bio-protocol 7(18): e2558. DOI: 10.21769/BioProtoc.2558.
Category
Immunology > Immune cell differentiation > MDSC
Cell Biology > Cell isolation and culture > Cell differentiation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link