- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Snapshots of the Signaling Complex DesK:DesR in Different Functional States Using Rational Mutagenesis and X-ray Crystallography
Published: Vol 7, Iss 16, Aug 20, 2017 DOI: 10.21769/BioProtoc.2510 Views: 8219
Reviewed by: Arsalan DaudiYann Simon GallotQiangjun Zhou

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Substituted Cysteine Accessibility Method for Topology and Activity Studies of Membrane Enzymes Forming Thioester Acyl Intermediates in Bacteria
Sébastien Gélis-Jeanvoine and Nienke Buddelmeijer
Nov 5, 2015 8791 Views

Aggregation Prevention Assay for Chaperone Activity of Proteins Using Spectroflurometry
Manish Bhuwan [...] Seyed E. Hasnain
Jan 20, 2017 12691 Views

Use of the Fluorescent Dye Thioflavin T to Track Amyloid Structures in the Pathogenic Yeast Candida albicans
Thierry Mourer [...] Sophie Bachellier-Bassi
Feb 5, 2024 2343 Views
Abstract
We have developed protocols to generate site-specific variants of the histidine-kinase DesK and its cognate response regulator DesR, conducive to trapping different signaling states of the proteins. Co-expression of both partners in E. coli, ensuring an excess of the regulator, was essential for soluble production of the DesK:DesR complexes and further purification. The 3D structures of the complex trapped in the phosphotransferase and in the phosphatase reaction steps, were solved by X-ray crystallography using molecular replacement. The solution was not trivial, and we found that in silico-generated models used as search probes, were instrumental to succeeding in placing a large portion of the complex in the asymmetric unit. Electron density maps were then clear enough to allow for manual model building attaining complete atomic models. These methods contribute to tackling a major challenge in the bacterial signaling field, namely obtaining stable kinase:regulator complexes, in distinct conformational states, amenable for high-resolution crystallographic studies.
Keywords: Signaling proteinsBackground
Structural information about bacterial signaling complexes, especially of two-component systems (TCSs), is still scarce (Casino et al., 2009; Gao and Stock, 2009). TCSs comprise a sensory histidine-kinase (HK) and a response regulator (RR) partner, present in almost all bacteria, they allow the cells to perceive the environment and to react accordingly through adaptive responses. Structural information is even more limited when it comes to TCS complexes adopting different functional states, despite the importance of such switching mechanism in signal transmission (Trajtenberg et al., 2016). We have studied the DesK-DesR pathway (de Mendoza, 2014), a TCS from Bacillus subtilis involved in regulating the cell membrane composition in adaptation to cues that reduce the bilayer’s fluidity, such as cold shock.
The protocols we have developed were aimed at overcoming major technical bottlenecks, encompassing complex purification, crystallization and X-ray structure determination. Most of these hurdles likely arise from the intrinsic flexibility and heterogeneity that characterize TCS proteins. With the purpose of trapping the DesK:DesR complex in defined signaling steps, it is useful to recall some details based on previous findings from our laboratory. The protocols have been developed to work with DesKC, a truncated DesK variant comprising the entire cytoplasmic region of DesK, without the trans-membrane sensory domain, which is catalytically competent to phosphotransfer to DesR, as well as to dephosphorylate P~DesR (Albanesi et al., 2004). As for the response regulator partner, DesR, we have chosen to use a truncated form, including the entire receiver domain (REC), competent for all DesK-mediated phosphotransfer reactions (Trajtenberg et al., 2014), but lacking the C-terminal DNA-binding domain, and thus minimizing potential inter-domain flexibility issues.
In order to trap the DesKC:DesR complex in the phosphotransfer step of the signaling pathway, we chose to use the phosphomimetic point mutant DesKC-His188Glu. This variant, when not bound to DesR, adopts a structural conformation very similar to the phosphorylated form of wild-type DesKC (Albanesi et al., 2009), hence an attractive template to mimic the phosphorylated HK just prior to the transfer reaction, also avoiding effective transfer to take place.
On the other hand, in order to trap the DesKC:DesR complex in the dephosphorylation step, previous work was instrumental by uncovering a switch mechanism of DesK, swapping between ‘active’ (kinase-on/phosphatase-off) and ‘inactive’ (phosphatase-on/kinase-off traits) states of the kinase (Albanesi et al., 2009). Briefly, the conformational transition of DesK from its kinase-active to the inhibited form, implicates the assembly of a coiled-coil structure within the central Dimerization and His-phosphotransfer (DHp) domain, a coiled-coil that is otherwise ‘broken’ when the kinase is active. The DHp, an all-helical domain, connects the trans-membrane sensor with the Catalytic ATP-binding (CA) domains, hence the identified DHp’s conformational switching plays a key role in signal transmission through long-range allosteric rearrangements. Such mechanistic insights later led to constructing a coiled-coil hyper-stabilized variant (DesKSTA) (Saita et al., 2015), harboring point-mutations at key positions (Ser150Ile, Ser153Leu and Arg157Ile) that stabilize a phosphatase-constitutive form (Saita et al., 2015). The corresponding soluble construct, with the trans-membrane domain truncated (DesKCSTAB), indeed displays a phosphatase-trapped 3D structure (Trajtenberg et al., 2016). DesKCSTAB was used to trap the DesKC:DesR complex in the dephosphorylation step, as described in this protocol.
Materials and Reagents
- P2, P200 and P1000 micropipette tips, autoclaved (Gilson, catalog numbers: F161630 , F161930 and F161670 )
- 1.5 ml Eppendorf tubes (Eppendorf, catalog number: 022364111 )
- 15 and 50 ml Falcon tubes (Corning, catalog numbers: 352097 and 352098 )
- Minisart 0.45 µm syringe filter (Sartorius, catalog number: 16555-K )
- 96-Well Clear V-Bottom 2 ml polypropylene deep well plate (Corning, catalog number: 3960 )
- Minisart 0.22 µm syringe filter (Sartorius, catalog number: 16532-K )
- 1 L polypropylene bottles (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 3140-1002 )
- SnakeSkin Dialysis Tubing 3.5K MWCO, 35 mm Dry I.D., 35 feet (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 88244 )
- Vivaspin 6 ml 10,000 MWCO centrifugal concentrator devices (Sartorius, catalog number VS0601 )
- Vivaspin 20 ml 10,000 MWCO centrifugal concentrator devices (Sartorius, catalog number VS2001 )
- Linbro 24-well plates (MP Biomedicals, catalog number: CPL-101 )
- Cryo-loops (HAMPTON RESEARCH, catalog number: HR4-955 )
- Cover slide
- Escherichia coli BL21 (DE3) and TOP10F’ strains from stocks stored at -80 °C
- pACYCDuet-1 (Novagen) and pQE80L (QIAGEN) plasmids
- Tobacco etch virus (TEV) protease (3 mg/ml stock solution, in-house preparation)
- Ultra-pure water (> 18 MΩ) filtered with 0.22 µm Express Plus filters (EMD Millipore, catalog number: SCGPT05RE )
- Ethanol 95% (Industrial Uruguayan Drugstore)
- Chloramphenicol (Sigma-Aldrich, catalog number: C0378 , 17 mg/ml stock solution, stored at -20 °C)
- Ampicillin (Sigma-Aldrich, catalog number: A9518 , 100 mg/ml stock solution, stored at -20 °C)
- Magnesium sulfate (MgSO4) (Sigma-Aldrich, catalog number: M7506 )
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Euromedex, catalog number: EU0008-B , 1 M stock solution, stored at -20 °C)
- Lysozyme (Sigma-Aldrich, catalog number: L6876 , 100 mg/ml stock solution)
- Triton X-100 (Sigma-Aldrich, catalog number: T9284 )
- Zinc chloride (ZnCl2) (Sigma-Aldrich, catalog number: 229997 )
- β-Mercaptoethanol (Sigma-Aldrich, catalog number: M6250 )
- Acrylamide/Bis-acrylamide 30% solution (Sigma-Aldrich, catalog number: A3574 )
- DNA Ladder GeneRuler 100 bp Plus (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: SM0321 )
- Oligonucleotides for mutagenesis and PCR amplifications (IDT DNA Technologies)
- Phusion High Fidelity DNA polymerase (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: F530S )
- Sodium dodecyl sulfate (Sigma-Aldrich, catalog number: L5750 )
- Ammonium persulfate (Sigma-Aldrich, catalog number: 248614 )
- N,N,N’,N’-Tetramethylethylenediamine (Sigma-Aldrich, catalog number: T9281 )
- Color Protein Ladder Prestained Broad Range (New England Biolabs, catalog number: P7712S )
- Precision Plus Protein Standard All Blue (Bio-Rad Laboratories, catalog number: 1610373 )
- Brilliant Blue R (Sigma-Aldrich, catalog number: B0149 )
- Lithium potassium acetyl phosphate (Sigma-Aldrich, catalog number: A0262 )
- Adenosine 5’-triphosphate (ATP) disodium salt hydrate (Sigma-Aldrich, catalog number: A1852 )
- β,γ-Methyleneadenosine 5’-triphosphate (AMP-PCP) disodium salt (Sigma-Aldrich, catalog number: M7510 )
- Trizma base (Sigma-Aldrich, catalog number: T1503 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: 31434 )
- Magnesium chloride hexahydrate (MgCl2•6H2O) (Sigma-Aldrich, catalog number: 13152 )
- Polyethylene glycol (PEG) 600 (Sigma-Aldrich, catalog number: 87333 )
- MES (Sigma-Aldrich, catalog number: M8250 )
- Magnesium sulfate (MgSO4)
- Glycerol (AppliChem, catalog number: 131339 )
- Polyethylene glycol (PEG) 4000 (Sigma-Aldrich, catalog number: 95904 )
- Lithium sulfate (Li2SO4) (Sigma-Aldrich, catalog number: 203653 )
- Beryllium chloride (BeCl2) (Sigma-Aldrich, catalog number: 201197 )
Note: This product has been discontinued. - Sodium fluoride (NaF) (Sigma-Aldrich, catalog number: 71519 )
- Liquid nitrogen
- Polyethylene glycol (PEG) 3350 (Sigma-Aldrich, catalog number: 202444 )
- Tri-potassium citrate (Sigma-Aldrich, catalog number: P1722 )
- Yeast extract (Sigma-Aldrich, catalog number: Y1625 )
- Tryptone plus (Sigma-Aldrich, catalog number: 61044 )
- Agar (Sigma-Aldrich, catalog number: A9799 )
- EDTA-free protease inhibitor cocktail tablets (Roche Diagnostics, catalog number: 11873580001 )
- Imidazole (Merck, catalog number: 104716 )
- Dithiothreitol (Soltec Ventures, catalog number: M112 )
- Hydrochloric acid solution (Sigma-Aldrich, catalog number: 13-1683 )
- LB medium (see Recipes)
- 2x YT culture medium (see Recipes)
- LB agar plates (see Recipes)
- Lysis buffer (see Recipes)
- Immobilized Metal Affinity Chromatography (IMAC) binding and washing buffer (see Recipes)
- IMAC elution buffer (see Recipes)
- Dialysis buffer (see Recipes)
- Size Exclusion Chromatography buffer for the phosphatase complex (SEC-P buffer)
- Size Exclusion Chromatography buffer for the phosphotransferase complex (SEC-PT buffer)
Equipment
- Pipetman P2 single channel pipette (Gilson, catalog number: F144801 )
- Pipetman P20 single channel pipette (Gilson, catalog number: F123600 )
- Pipetman P200 single channel pipette (Gilson, catalog number: F123601 )
- Pipetman P1000 single channel pipette (Gilson, catalog number: F123602 )
- 250 ml Erlenmeyer flasks (Marienfeld-Superior, catalog number: 4110207 )
- 5 L Erlenmeyer flasks (Marienfeld-Superior, catalog number: 4110217 )
- HisTrap immobilized metal affinity chromatography (IMAC) 5 ml column with Zn-NTA resin (GE Healthcare, catalog number: 17-5248-01 . In-house preparation)
- Superdex S75 26/600 (GE Healthcare, catalog number: 28989334 )
- Stirring magnet (Sigma-Aldrich)
- Thermomixer C (Eppendorf, model: ThermoMixer® C , catalog number: 4053-8223)
- Digital Ultrasonics Sonifier S-450 cell disruptor/homogenizer (Emerson, Branson Ultrasonics, model: S-450 )
- Multitron Standard incubation shaker (Infors HT)
- Minispin centrifuge (Eppendorf, model: MiniSpin® , catalog number: 5452000018)
- Refrigerated 5424R Centrifuge (Eppendorf, model: 5424 R , catalog number: 36-102-3795)
- Refrigerated 5810R Centrifuge (Eppendorf, model: 5810 R , catalog number: 5811000010)
- Refrigerated Sorvall Lynx4000 centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: Sorvall Lynx4000 , catalog number: 75006580)
- Minipuls 3 peristaltic pump (Gilson, catalog number: F117604 )
- Chromatography instrument ÄKTA purifier (GE Healthcare, model: ÄKTAxpress , catalog number: 18664501)
- EPS-301 power supply (GE Healthcare, model: EPS 301, catalog number: 18-1130-01 )
- Cary 50 Bio UV-visible spectrophotometer (Varian, model: Cary® 50 )
- Alchemist DT (Rigaku, model: Alchemist DT )
- X-ray generator MicroMax-007HF (Rigaku, model: MicroMax-007 HF )
- Multilayer X-ray mirrors Varimax-HF (Rigaku, model: VariMax HF )
- Image plate area detector MAR345® (marXperts, model: mar345 )
- SZX16 microscope (Olympus, model: SZX16 )
- EVOLT E-330 digital camera (Olympus, model: E-330 )
- LG-PS2 light source (Olympus, model: LG-PS2 )
- Linux Computer workstation with OS Centos 7.0
Software
- Produced diffraction datasets:
- Complex DesKC:DesR-REC in phosphotransferase state, low Mg2+:
http://dx.doi.org/10.15785/SBGRID/399 - Complex DesKC:DesR-REC in phosphotransferase state, high Mg2+:
http://dx.doi.org/10.15785/SBGRID/401 - Complex DesKC:DesR-REC in phosphotransferase state, high Mg2+ and BeF3-:
http://dx.doi.org/10.15785/SBGRID/408 - Complex DesKC:DesR-REC in phosphatase state:
http://dx.doi.org/10.15785/SBGRID/400
- Complex DesKC:DesR-REC in phosphotransferase state, low Mg2+:
- Produced atomic coordinate models:
- Complex DesKC:DesR-REC in phosphotransferase state, low Mg2+:
http://www.rcsb.org/pdb/explore/explore.do?structureId=5IUJ - Complex DesKC:DesR-REC in phosphotransferase state, high Mg2+:
http://www.rcsb.org/pdb/explore/explore.do?structureId=5IUK - Complex DesKC:DesR-REC in phosphotransferase state, high Mg2+ and BeF3-:
http://www.rcsb.org/pdb/explore/explore.do?structureId=5IUL - Complex DesKC:DesR-REC in phosphatase state:
http://www.rcsb.org/pdb/explore/explore.do?structureId=5IUN
- Complex DesKC:DesR-REC in phosphotransferase state, low Mg2+:
- Computational crystallography software utilized:
- autoPROC (https://www.globalphasing.com/autoproc)
- BUSTER (https://www.globalphasing.com/buster)
- CCP4 (http://www.ccp4.ac.uk)
- Phaser (http://www.phaser.cimr.cam.ac.uk)
- Coot (https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot)
- MolProbity (http://molprobity.biochem.duke.edu)
- PyMol (https://www.pymol.org)
Procedure
- The use of DesKC mutants to stabilize the histidine-kinase either in its phosphatase or its phosphotransferase state
- The plasmid pACYC-DesKCH188E:DesRREC encoding for the phosphomimetic DesKC variant and for the receiver domain of DesR (DesRREC), both fused to a His-tag and a TEV protease site to cleave the tag, was already available (Trajtenberg et al., 2014). This plasmid pACYC-DesKCH188E:DesRREC is thus used to co-express the following two proteins:
- DesKCH188E:
MGSSHHHHHHGIHMENLYFQGRKERERLEEKLEDANERIAELVKLEERQRIARDLEDTLGQKLSLIGLKSDLARKLIYKDPEQAARELKSVQQTARTSLNEVRKIVSSMKGIRLKDELINIKQILEAADIMFIYEEEKWPENISLLNENILSMCLKEAVTNVVKHSQAKTCRVDIQQLWKEVVITVSDDGTFKGEENSFSKGHGLLGMRERLEFANGSLHIDTENGTKLTMAIPNNSK
Theoretical MW = 27.3 kDa (24.9 after TEV-cleavage)
Theoretical absorbance (280 nm) of a 1 mg/ml solution after TEV-cleavage = 0.56 - DesRREC:
MRGSHHHHHHGSGSENLYFQGSGSMISIFIAEDQQMLLGALGSLLNLEDDMEVVGKGTTGQDAVDFVKKRQPDVCIMDIEMPGKTGLEAAEELKDTGCKIIILTTFARPGYFQRAIKAGVKGYLLKDSPSEELANAIRSVMNGKRIYAPELMEDLYSEA
Theoretical MW = 17.4 kDa (15.2 after TEV-cleavage)
Theoretical absorbance (280 nm) of a 1 mg/ml solution after TEV-cleavage = 0.40
- DesKCH188E:
- Within the sequences listed in the previous point, His-tags are highlighted with blue fonts, in green, the TEV-recognition sites (cleaved proteins start at the final Gly, including it). Underlined in red, the phosphorylation sites: DesRREC bears the wild-type Asp, while DesKCH188E displays instead a Glu replacing the native His.
- The phosphatase-stabilized variant of DesKC, DesKCSTAB, is insoluble when expressed by itself in Escherichia coli. This difficulty is solved by co-expressing DesKCSTAB with DesRREC, resulting in excellent yields of both proteins. A co-expression plasmid is generated (pACYC-DesKCSTAB:DesRREC), by sub-cloning DesKCSTAB from pHPKS/Pxyl-desKSTA (Saita et al., 2015) into pACYC-DesKCH188E:DesRREC (Trajtenberg et al., 2014) using restriction-free cloning (Unger et al., 2010), using primers STAB_F (5’-CCTGTATTTTCAGGGATCCGGTATTATAAAACTTCGCAAG-3’) and STAB_R (5’-GTCAGACACTGTAATCACAACTTCCTTCCAG-3’). Both recombinant proteins encoded in pACYC-DesKCSTAB:DesRREC include a His-tag and a TEV protease cleavage site.
- pACYC-DesKCSTAB:DesRREC co-expresses the following two proteins:
- DesKCSTAB:
MGSSHHHHHHGSGSENLYFQGSGIIKLRKEIERLEEKLEDANERIAELVKLEERQRIARDLHDTLGQKLSLIGLKSDLARKLIYKDPEQAARELKSVQQTARTSLNEVRKIVSSMKGIRLKDELINIKQILEAADIMFIYEEEKWPENISLLNENILSMCLKEAVTNVVKHSQAKTCRVDIQQLWKEVVITVSDDGTFKGEENSFSKGHGLLGMRERLEFANGSLHIDTENGTKLTMAIPNNSK
Theoretical MW = 27.8 kDa (25.6 after TEV-cleavage)
Theoretical absorbance (280 nm) of a 1 mg/ml solution after TEV-cleavage = 0.55 - DesRREC:
MRGSHHHHHHGSGSENLYFQGSGSMISIFIAEDQQMLLGALGSLLNLEDDMEVVGKGTTGQDAVDFVKKRQPDVCIMDIEMPGKTGLEAAEELKDTGCKIIILTTFARPGYFQRAIKAGVKGYLLKDSPSEELANAIRSVMNGKRIYAPELMEDLYSEA
Theoretical MW = 17.4 kDa (15.2 after TEV-cleavage)
Theoretical absorbance (280 nm) of a 1 mg/ml solution after TEV-cleavage = 0.40
Note: the theoretical absorbance (280 nm) of a 1 mg/ml solution of the DesKC:DesR protein complex is approximately 0.5, which corresponds to both protein sequences together.
- DesKCSTAB:
- Within the sequences listed in the previous point, His-tags are highlighted with blue fonts, in green, the TEV-recognition sites (cleaved proteins start at the final Gly, including it). Underlined in orange, the three stabilizing substitutions within the coiled-coil motif of DesK’s DHp domain. Underlined in red, the phosphorylation sites.
- The plasmid pACYC-DesKCH188E:DesRREC encoding for the phosphomimetic DesKC variant and for the receiver domain of DesR (DesRREC), both fused to a His-tag and a TEV protease site to cleave the tag, was already available (Trajtenberg et al., 2014). This plasmid pACYC-DesKCH188E:DesRREC is thus used to co-express the following two proteins:
- Over-expression of DesKC:DesR complexes
- The co-expression plasmid pACYC-DesKCSTAB:DesRREC (and similarly for pACYC-DesKCH188E:DesRREC), is transformed into 25 µl of competent Escherichia coli BL21 (DE3) cells (approximately 108 cells/ml) using heat shock for 2 min at 42 °C. A pQE80L plasmid including the DesRREC sequence (Trajtenberg et al., 2014) is co-transformed to produce a stoichiometric excess of DesRREC in the cells. 100 μl of transformed cells are spread on LB agar plates containing 100 μg/ml ampicillin and 17 μg/ml chloramphenicol, and grown overnight at 37 °C.
- Between 5 and 10 single colonies are picked and inoculated in 3 ml LB culture media (to be used as pre-cultures in the next step), 100 μg/ml ampicillin and 17 μg/ml chloramphenicol, and further grown at 37 °C for 16 h with agitation (220 rpm).
- Pre-cultures are inoculated 1/500 in 250 ml Erlenmeyer flasks containing 50 ml LB media, 100 μg/ml ampicillin and 17 μg/ml chloramphenicol, and further grown at 37 °C for 16 h with agitation (220 rpm).
- Larger-scale cultures are launched using three 5 L Erlenmeyer flasks, each containing 1 L 2x YT culture media, 2 mM magnesium sulfate, 100 μg/ml ampicillin, 17 μg/ml chloramphenicol and a 1/100 inoculum of pre-cultured bacteria. Cultures are grown at 37 °C with agitation (220 rpm) until absorbance at wavelength 660 nm (Abs660 nm) reaches 0.7-1 (approximately 3 h). Cultures are quickly shifted to a shaker that has been pre-equilibrated at 20 °C, and further growth is continued for 30 min with agitation (220 rpm).
- IPTG is added (0.5 mM final concentration) into the cultures. Induction of over-expression is achieved at 30 °C for 4 h (in the case of DesKCSTAB:DesRREC) or at 20 °C for 16 h (for DesKCH188E:DesRREC), with agitation (220 rpm) in both cases.
- Cells are harvested by centrifugation at 4,600 x g for 20 min at 4 °C in 1 L polypropylene bottles.
- Pellets are thoroughly resuspended in lysis buffer (see Recipes) (5 ml per g wet weight pellet) in 50 ml Falcon tubes.
- Lysozyme (0.5 mg/ml final concentration) and Triton X-100 (1% vol/vol final concentration) are added to resuspended pellets, and incubated at room temperature for 30 min, until the extract is viscous due to DNA release. Extracts are then transferred to -80 °C and stored for at least 2 h.
- Frozen extracts are thawed in a water bath at 37 °C. Thawed extracts are sonicated with pulses of 1 sec (set to 30% amplitude) and resting intervals of 3 sec, completing a total timespan of 4 min. This operation is repeated 3-4 times, until complete reduction of viscosity. During sonication the Falcon tube is kept refrigerated with ice. This is considered the total extract, and 40 μl are stored in electrophoresis sample buffer with β-mercaptoethanol (SBβ) for later SDS-PAGE analyses.
- Total extracts are centrifuged at 12,000 x g and 4 °C for 45 min. This step separates soluble species (supernatant fraction) from insoluble ones (pellet) such as inclusion bodies, non-lysed cells, insoluble proteins, etc. The supernatants are filtered through a 0.45 μm syringe filter to a new Falcon tube, and imidazole added to 40 mM final concentration (to avoid nonspecific binding to the column during IMAC). 40 μl of supernatants are stored in SBβ for SDS-PAGE analysis. Supernatants are hereafter used as the source of soluble DesKCSTAB (or DesKCH188E) and DesRREC, for structural studies.
- The co-expression plasmid pACYC-DesKCSTAB:DesRREC (and similarly for pACYC-DesKCH188E:DesRREC), is transformed into 25 µl of competent Escherichia coli BL21 (DE3) cells (approximately 108 cells/ml) using heat shock for 2 min at 42 °C. A pQE80L plasmid including the DesRREC sequence (Trajtenberg et al., 2014) is co-transformed to produce a stoichiometric excess of DesRREC in the cells. 100 μl of transformed cells are spread on LB agar plates containing 100 μg/ml ampicillin and 17 μg/ml chloramphenicol, and grown overnight at 37 °C.
- Purification of DesKC:DesR complexes
- Zinc is used as the immobilized metal, during the first purification step by IMAC. Nickel is avoided to preclude cysteine oxidation, which previously hampered crystallization due to resulting heterogeneity in DesR samples. The IMAC column is connected to the ÄKTA purifier and pre-equilibrated with binding buffer (see Recipes) (10 column volumes or CV). Samples are injected at a flow rate of 5 ml/min, and target proteins are bound to the column through their N-terminal His-tags.
- IMAC washing is achieved with 15-20 CV binding buffer, until a stable Abs280 baseline is obtained. Aliquots of the flow-through material are stored in SBβ for SDS-PAGE analysis, to monitor for potential column saturation.
- IMAC elution is achieved with a linear 0-100% gradient of elution buffer (see Recipes) (30 CV), at 5 ml/min. Absorbance at wavelength 280 nm is used to monitor protein elution peaks. DesKCSTAB, DesKCH188E and DesRREC typically elute at approximately 65 mM imidazole and are collected in 96-deep-well plates. 40 μl of eluted peaks are stored in SBβ for SDS-PAGE analysis.
- The elution peaks of selected proteins are pooled and incubated with TEV protease (1:40 w/w TEV/target ratio). Proteolysis is performed overnight, in dialysis bags. Dialyses are performed against 200-300 volumes of dialysis buffer (see Recipes) with gentle stirring.
- Samples are recovered from the dialysis bag, and filtered with 0.45 μm syringe filters. A second IMAC zinc column is used, attached to a peristaltic pump and pre-equilibrated with binding buffer (10 CV). Samples are injected into the column at a flow rate of 5 ml/min. The flow-through is now carefully collected. The TEV protease itself includes an N-terminal His-tag, hence typically excluded from the flow-through. 20 CV of elution buffer is applied to the column to elute TEV (and potentially non-digested target protein, normally absent if the procedure worked correctly) to be monitored by SDS-PAGE.
- The second IMAC flow-through sample is concentrated by ultra-filtration in Vivaspin centrifuge devices at 6,300 x g and 15 °C for 20 min to a final volume of 10 ml. This is immediately filtered through a 0.22 μm syringe filter, and injected into a size exclusion chromatography (SEC) column Superdex S75 26/600, at a flow rate of 1 ml/min. The SEC column is previously equilibrated with 2 CV SEC buffer (see Recipes).
- The SEC is eluted isocratically at 1 ml/min with SEC-P buffer in the case of the DesKCSTAB:DesRREC (phosphatase) complex, or with SEC-PT buffer in the case of the DesKCH188E:DesRREC (phosphotransferase) complex. Eluted fractions are collected in 96-deep-well plates. Protein elution is monitored with 280 nm wavelength absorbance (Figures 1A and 2A). 40 μl of eluted peaks are stored in SBβ for SDS-PAGE analysis.
The calculated molecular weight (MW) of the complex is approximately 65 kDa taking into account that DesKC is a homo-dimer, and that one monomer of DesRREC could be binding to DesKC. If instead the DesKC:DesRREC ratio is 2:2, the MW is anticipated to increase to ~80 kDa. According to the calibration curves, the SEC chromatograms reveal a MW of 53.5 kDa in the case of DesKCSTAB:DesRREC, and 44.9 kDa for DesKCH188E:DesRREC. The difference with the theoretical MW figures is likely due to tertiary and quaternary structure features deviating from the ideal assumptions for globular species and their hydrodynamic radii. That these SEC peaks correspond indeed to the targeted DesKC:DesR complexes is afterwards confirmed by mass spectrometry analyses and then 3D structure determination. The final yields approximated 12 mg protein per L of cell culture (or 1-2 mg protein per g of wet weight pellet), for both complexes.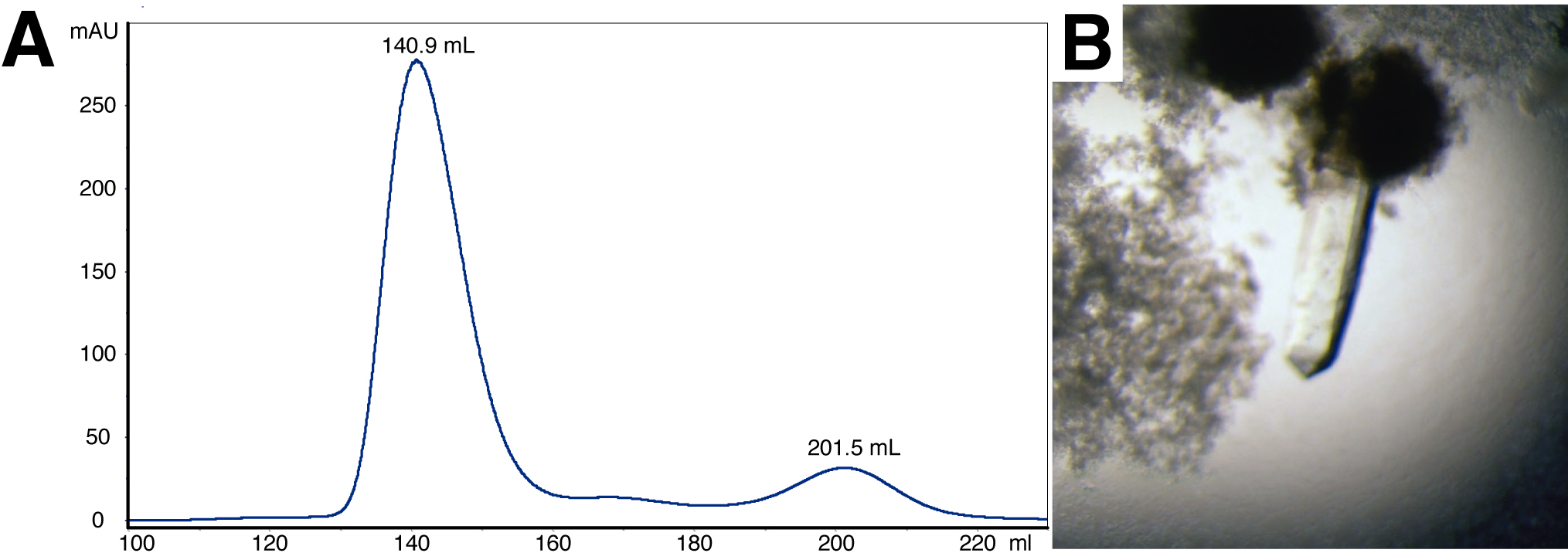
Figure 1. Size-exclusion chromatography and crystallization of the DesKCSTAB:DesRREC complex. A. Elution profile of the DesKCSTAB:DesRREC protein complex in a Superdex S75 pg 26/600 size exclusion chromatography column. The peak at 140.9 ml corresponds to the complex, according to SDS-PAGE analysis. The peak at 201.5 ml corresponds to monomeric DesRREC in excess. B. Trigonal crystal of the DesKCSTAB:DesRREC complex.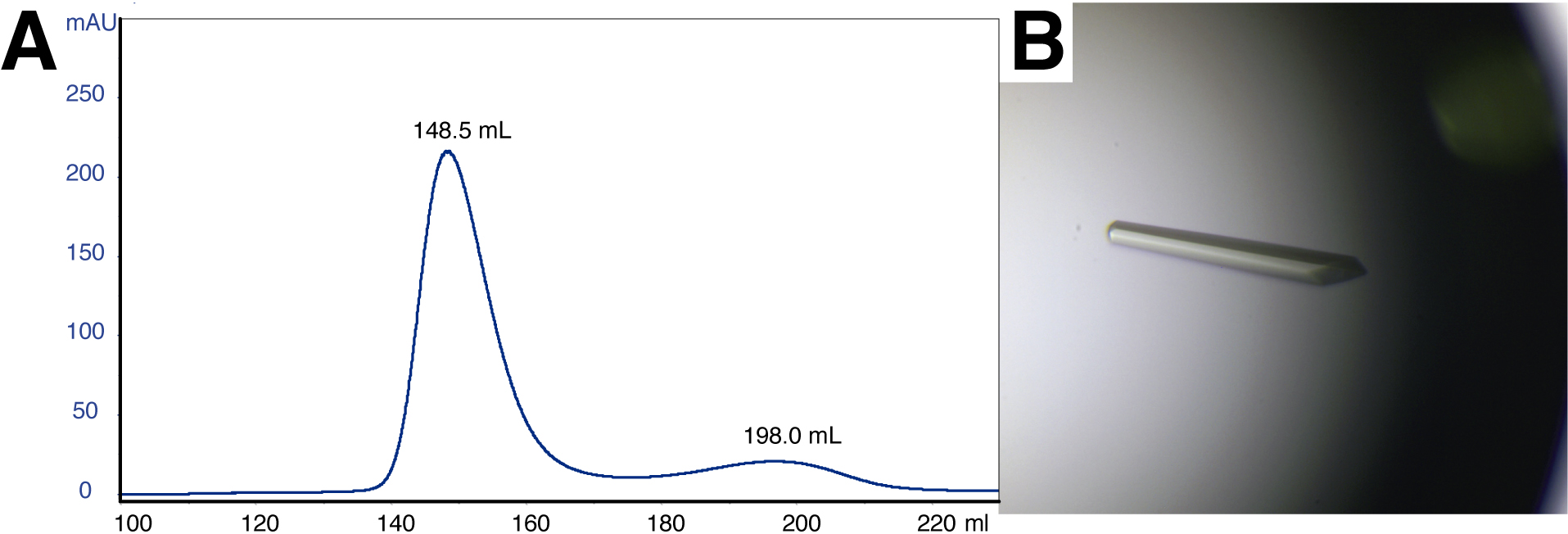
Figure 2. Size-exclusion chromatography and crystallization of the DesKCH188E:DesRREC complex. A. Elution profile of the DesKCH188E:DesRREC protein complex in a Superdex S75 pg 26/600 size exclusion chromatography column. The peak at 148.5 ml corresponds to the complex, according to SDS-PAGE analysis. The peak at 198.0 ml corresponds to monomeric DesRREC in excess. B. Monoclinic crystal of the DesKCH188E:DesRREC complex.
Selected elution peaks from SEC are pooled and concentrated with Vivaspin centrifuge devices at 6,300 x g and 15 °C for 20 min. Final concentrations of 16 mg/ml for the DesKCSTAB:DesRREC complex, and 19 mg/ml for the DesKCH188E:DesRREC complex, are typically achieved. Protein concentration is determined by UV spectrophotometry at 280 nm and calculated extinction coefficients are derived from ProtParam (ExPASy, SIB Bioinformatics Resource Portal: http://www.expasy.org/tools). Proteins are stored in 25, 50 and 100 µl aliquots at -80 °C. - Prior to crystallization, the purity and integrity of samples are checked by SDS-PAGE. Samples in SBβ are heated at 100 °C for 5 min and separated by electrophoresis in 12% polyacrylamide gels ran at 200 V.
- Zinc is used as the immobilized metal, during the first purification step by IMAC. Nickel is avoided to preclude cysteine oxidation, which previously hampered crystallization due to resulting heterogeneity in DesR samples. The IMAC column is connected to the ÄKTA purifier and pre-equilibrated with binding buffer (see Recipes) (10 column volumes or CV). Samples are injected at a flow rate of 5 ml/min, and target proteins are bound to the column through their N-terminal His-tags.
- Crystallization of DesKC:DesR complexes
- DI: The DesKCSTAB:DesRREC (phosphatase) complex
- The protein stock solution of pure DesKCSTAB:DesRREC complex is prepared according to a recipe where the order in the addition of the reagents is critical. First MIXA is obtained by combining 7.6 μl 100 mM AMP-PCP, 3.8 μl 1 M Tris pH 8.5 and 44.6 μl buffer SEC-P. Then 95 μl of DesKCSTAB:DesRREC complex (at ~16 mg/ml in buffer SEC-P) is added to the 56 μl of MIXA. The tube is centrifuged at 16,000 x g and 4 °C for 10 min and the supernatant used for further manipulations. In this manner ~150 µl of DesKCSTAB:DesRREC complex stock solution (at ~10 mg/ml final concentration) is obtained, containing approximately 5 mM AMP-PCP, 43.7 mM Tris pH 8.5, 462.2 mM NaCl and 9.25 mM MgCl2.
- Microseeding is used to speed up the process of crystallogenesis: 2 μl are drawn from drops containing previously grown crystals, and these are crushed by vigorous pipetting with P2 micropipette tips. This is used as the source of microseeds, adding 50 μl of 30% (v/v) PEG 600, 0.1 M MES pH 6, 0.15 M MgSO4 and 5% (v/v) glycerol. This seed stock solution is diluted 1/400 into an additive solution previously prepared containing 27% (v/v) PEG 600, 0.1 M MES pH 6, 0.15 M MgSO4 and 5% (v/v) glycerol.
- Hanging drop crystallizations are done at 20 °C in Linbro plates. The crystallization drops contain 0.8 μl of mother liquor (30% [w/v] PEG 4000, 0.1 M Tris-HCl pH 8.5, 0.2 M Li2SO4), 2 μl of stock protein solution, and 1.2 μl of additive solution with seeds. Three drops are set on a cover slide to seal each reservoir well. Crystals typically appear in 3-4 days, growing to suitable sizes (0.5 µm) in 10 days (Figure 1B).
- Crystals are cryo-protected by slowly adding 4 μl of cryo-protection solution: 32% (w/v) PEG 4000, 0.1 M Tris-HCl pH 8, 0.2 M Li2SO4, 20 mM MgCl2, 18 mM BeF3-, 5 mM AMP-PCP and 15% glycerol. BeF3- is prepared by mixing 50 μl of a 0.9 M stock solution of NaF with 9 μl of a 1 M stock solution of BeCl2, rendering a stock solution of 152 mM BeF3-. Be is extremely toxic; all solutions containing it are handled with particular caution and wastes treated appropriately, according to safety rules. Finally, crystals are briefly soaked in 100% cryoprotection solution, fished out of the drops using cryo-loops of approximately the same size as the selected specimen, flash-frozen in liquid nitrogen and stored in cryo-vials under liquid nitrogen for further use.
- The protein stock solution of pure DesKCSTAB:DesRREC complex is prepared according to a recipe where the order in the addition of the reagents is critical. First MIXA is obtained by combining 7.6 μl 100 mM AMP-PCP, 3.8 μl 1 M Tris pH 8.5 and 44.6 μl buffer SEC-P. Then 95 μl of DesKCSTAB:DesRREC complex (at ~16 mg/ml in buffer SEC-P) is added to the 56 μl of MIXA. The tube is centrifuged at 16,000 x g and 4 °C for 10 min and the supernatant used for further manipulations. In this manner ~150 µl of DesKCSTAB:DesRREC complex stock solution (at ~10 mg/ml final concentration) is obtained, containing approximately 5 mM AMP-PCP, 43.7 mM Tris pH 8.5, 462.2 mM NaCl and 9.25 mM MgCl2.
- DII: The DesKCH188E:DesRREC (phosphotransferase) complex
- The protein stock solution of pure DesKCH188E:DesRREC complex is prepared according to a recipe where the order in the addition of the reagents is critical. First MIXA is obtained by combining 5.5 μl 100 mM AMP-PCP, 2.75 μl 1 M Tris pH 8.5, 2.2 μl 1 M MgCl2 and 43.35 μl buffer SEC-PT. Then 48 μl of DesKCH188E:DesRREC complex (at ~19 mg/ml in buffer SEC-PT) are added to 8.2 μl DesRREC (at 30 mg/ml in buffer SEC-PT) and the 53.8 μl of MIXA. The tube is centrifuged at 16,000 x g and 4 °C for 10 min and the supernatant used for further manipulations. In this manner ~110 µl of DesKCH188E:DesRREC complex stock solution (at ~8.3 mg/ml final concentration) is obtained, containing approximately 5 mM AMP-PCP, 43.1 mM Tris pH 8-8.5, 271.5 mM NaCl and 20 mM MgCl2.
- Hanging drop crystallizations are done at 20 °C in Linbro plates. The crystallization drops contain 2 μl of mother liquor (18% PEG 3350, 0.3 M tri-potassium citrate) and 2 μl of stock protein solution. Two drops are set on a cover slide to seal each reservoir well. Crystals appear typically in 5-6 days, growing to suitable sizes (0.5 µm) in 15 days (Figure 2B).
- Crystals are cryo-protected by quick soaking in 20% PEG 3350, 0.3 M tri-potassium citrate, 5 mM AMP-PCP, 25% glycerol, 20 or 150 mM MgCl2, and 0 or 5 mM BeF3-. Crystals are fished out of the drops using cryo-loops of approximately the same size as the selected specimen, flash-frozen in liquid nitrogen, and stored in cryo-vials under liquid nitrogen for further use.
- The protein stock solution of pure DesKCH188E:DesRREC complex is prepared according to a recipe where the order in the addition of the reagents is critical. First MIXA is obtained by combining 5.5 μl 100 mM AMP-PCP, 2.75 μl 1 M Tris pH 8.5, 2.2 μl 1 M MgCl2 and 43.35 μl buffer SEC-PT. Then 48 μl of DesKCH188E:DesRREC complex (at ~19 mg/ml in buffer SEC-PT) are added to 8.2 μl DesRREC (at 30 mg/ml in buffer SEC-PT) and the 53.8 μl of MIXA. The tube is centrifuged at 16,000 x g and 4 °C for 10 min and the supernatant used for further manipulations. In this manner ~110 µl of DesKCH188E:DesRREC complex stock solution (at ~8.3 mg/ml final concentration) is obtained, containing approximately 5 mM AMP-PCP, 43.1 mM Tris pH 8-8.5, 271.5 mM NaCl and 20 mM MgCl2.
- DI: The DesKCSTAB:DesRREC (phosphatase) complex
- X-ray diffraction data collection.
- Single crystal X-ray diffraction experiments are carried out with an in-house copper rotating-anode source (Protein Crystallography Facility, Institut Pasteur Montevideo), or with synchrotron radiation (Soleil, France).
- DesKCSTAB in complex with DesRREC (PDB Id 5IUN), crystallizes in the trigonal space group P3121. Crystals are measured in the synchrotron (Beamline Proxima I, Soleil, France), collecting 180 images with 1° oscillation range and 30 sec exposure time per image (Figure 3A). The raw data is deposited in the SBGrid Data Bank (DOI: 10.15785/SBGRID/400).
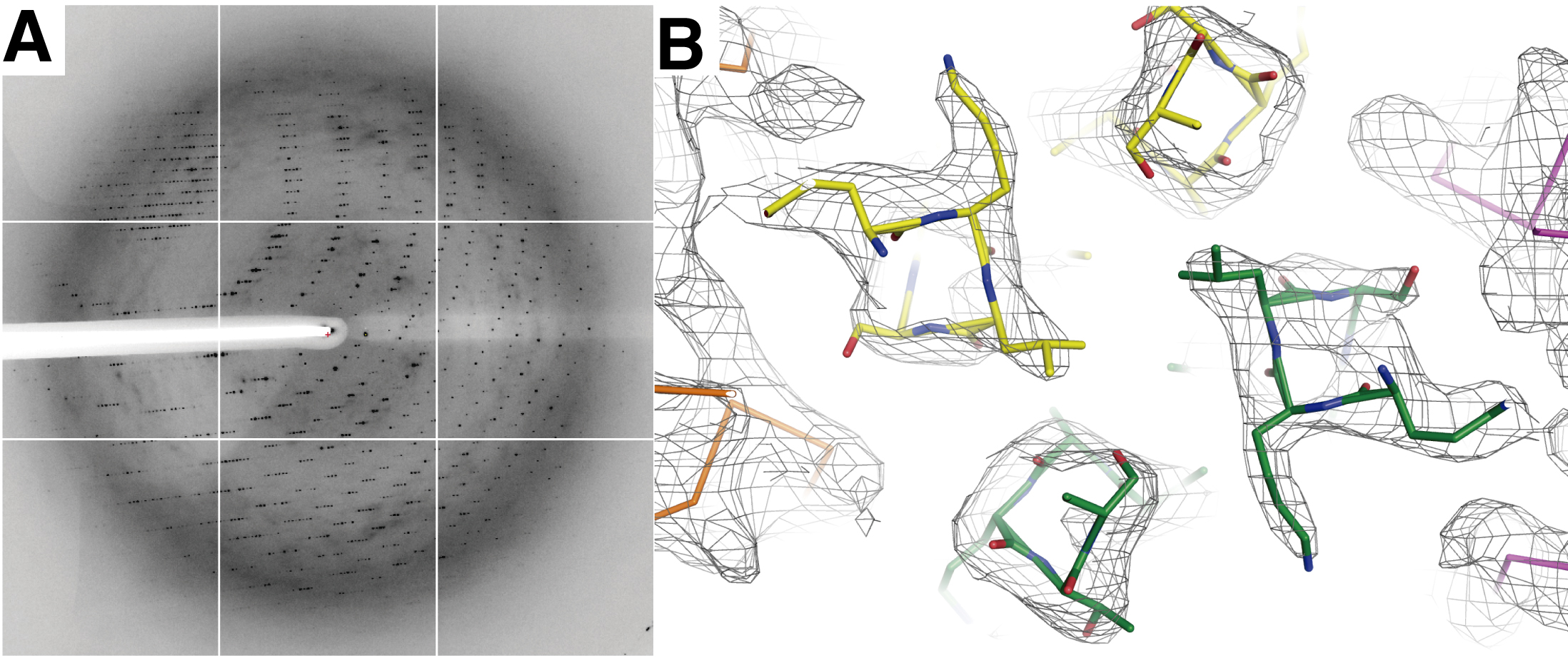
Figure 3. X-ray diffraction of DesKCSTAB:DesRREC crystals and resulting electron density map. A. Representative frame showing the X-ray diffraction from a single DesKCSTAB:DesRREC crystal (space group P3121). B. Zoom-in on the mid-sector of the DHp domain of the DesKCSTAB:DesRREC (phosphatase) complex, illustrated in stick representation (all atoms for the kinase, and only the Cα trace for the regulator). Oxygen atoms are colored red, nitrogens in blue, and carbons distinguished according to protein and chain: green and yellow, for the two chains in the DesKCSTAB dimer, and orange and magenta for the two bound DesRREC moieties. - DesKCH188E in complex with DesRREC with high Mg2+ and BeF3- (PDB Id 5IUL), is collected in a rotating anode X-ray generator (Protein Crystallography Facility, Institut Pasteur de Montevideo, Uruguay). 593 images are collected with 0.3° oscillation range and 10 min exposure per image (Figure 4A). The raw data is deposited in the SBGrid Data Bank (DOI: 10.15785/SBGRID/408).
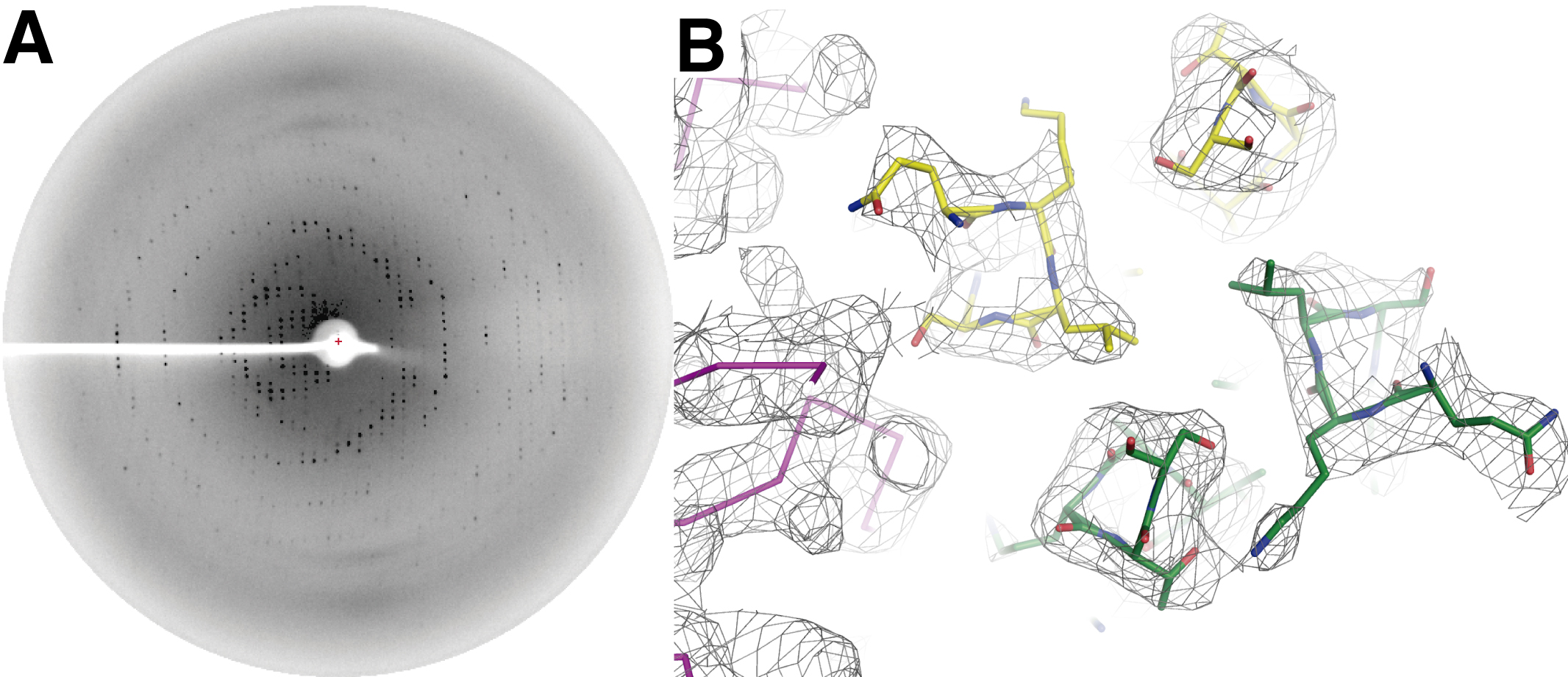
Figure 4. X-ray diffraction of DesKCH188E:DesRREC crystals and resulting electron density map. A. Representative frame showing the X-ray diffraction from a single DesKCH188E:DesRREC crystal (space group P21). B. Zoom-in on the mid-sector of the DHp domain of the DesKCH188E:DesRREC (phosphotransferase) complex, illustrated in stick representation (all atoms for the kinase, and only the Cα trace for the regulator). Oxygen atoms are colored red, nitrogens in blue, and carbons distinguished according to protein and chain: green and yellow, for the two chains in the DesKCH188E dimer, and magenta for the single asymmetrically bound DesRREC molecule.
DesKCH188E in complex with DesRREC with high Mg2+ (PDB Id 5IUK), crystallizes in the monoclinic space group P21. Crystals are measured in the synchrotron (Beamline Proxima I, Soleil, France), collecting two different sets of 1,000 frames each. The oscillation range is 0.2° and exposure time is 0.2 sec per image. The raw data is deposited in the SBGrid Data Bank (DOI: 10.15785/SBGRID/401). - DesKCH188E in complex with DesRREC with low Mg2+ (PDB Id 5IUJ), crystallizes in the monoclinic space group P21. Crystals are measured in the synchrotron (Beamline Proxima II, Soleil, France), collecting 110 images with 1° oscillation range and 30 sec exposure per image. The raw data is deposited in the SBGrid Data Bank (DOI: 10.15785/SBGRID/399).
- DesKCSTAB in complex with DesRREC (PDB Id 5IUN), crystallizes in the trigonal space group P3121. Crystals are measured in the synchrotron (Beamline Proxima I, Soleil, France), collecting 180 images with 1° oscillation range and 30 sec exposure time per image (Figure 3A). The raw data is deposited in the SBGrid Data Bank (DOI: 10.15785/SBGRID/400).
- Single crystal X-ray diffraction experiments are carried out with an in-house copper rotating-anode source (Protein Crystallography Facility, Institut Pasteur Montevideo), or with synchrotron radiation (Soleil, France).
Data analysis
- Data sets are processed using the automatic pipeline autoPROC (Vonrhein et al., 2011), which uses XDS (Kabsch, 2010) for indexing/integration, and Pointless/Aimless (Evans, 2006; Evans, 2011) for data reduction and scaling, with the following comments for each case:
- DesKCSTAB in complex with DesRREC (PDB Id 5IUN): the best processing statistics are achieved by integrating all images.
- DesKCH188E in complex with DesRREC with high Mg2+ (PDB Id 5IUK): the best processing strategy is to integrate and scale the first 800 frames from the first data set, merging it with frames 1-530 and 650-910 from the second set.
- DesKCH188E in complex with DesRREC with high Mg2+ and BeF3- (PDB Id 5IUL): all images are eventually integrated, but successful indexing is achieved by selecting frames 10-40 and 200-240 and using the 1,000 strongest reflections.
- DesKCH188E in complex with DesRREC with low Mg2+ (PDB Id 5IUJ): the best processing statistics are achieved by integrating all images.
- DesKCSTAB in complex with DesRREC (PDB Id 5IUN): the best processing statistics are achieved by integrating all images.
- To solve the structure of DesKCSTAB in complex with DesRREC (Figure 3B), molecular replacement is used, as previously reported (Trajtenberg et al., 2016). The search probe is a model of a DesKC:DesRREC complex generated in silico (Trajtenberg et al., 2014) by superposition of a distantly related complex from Termothoga maritima (PDB Id 3DGE) (Casino et al., 2009), and partial truncation of DesKC’s DHp domain, only keeping the invariant region (residues 190-230). It must be highlighted that the in silico modeling strategy produces hundreds of models (Trajtenberg et al., 2014). Molecular replacement is performed using Phaser (McCoy et al., 2007), searching for one copy of the in silico-generated DHp-DesRREC model of the complex, and repeating this automatically with hundreds of different initial candidates as search probes. Eventually only a handful of them are good to be placed in the asymmetric unit, giving clear signals in the rotation and the translation functions calculated with default settings. Starting with the first refinement cycles, the remaining domains and a second hemi-complex are clearly visible in the electron density maps, allowing for manual model building of the whole molecules using Coot (Emsley et al., 2010). The strategy of using in silico-generated DHp-DesRREC models as search probes is critical for molecular replacement to succeed, and the final refined model proves indeed that the selected probes were similar enough to allow for molecular replacement to succeed, providing with solutions within the radius of convergence for refinement procedures (Figure 5). Using instead crystallographic models of individual partners or domains instead is ineffective, likely due to insufficient scattering mass of small search probes and/or conformational differences between isolated vs. complexed proteins.
The structures corresponding to the phosphotransferase complex are also solved by molecular replacement with Phaser as before, using the DHp-DesRREC model as search probe, and then completing as described above, guided by the electron density maps. Once more, we refer the reader to our previous report for full details of crystallographic structure determination procedures (Trajtenberg et al., 2016).- Structure refinement is performed with Buster-TNT (Bricogne et al., 2009) using standard procedures, including non-crystallographic symmetry (NCS) restraints and Translation/Libration/Screw (TLS) descriptions in each model (documented in detail within the header of each atomic coordinates file available in the PDB).
- Structures are validated throughout the refinement and towards the end, using MolProbity tools (Chen et al., 2010).
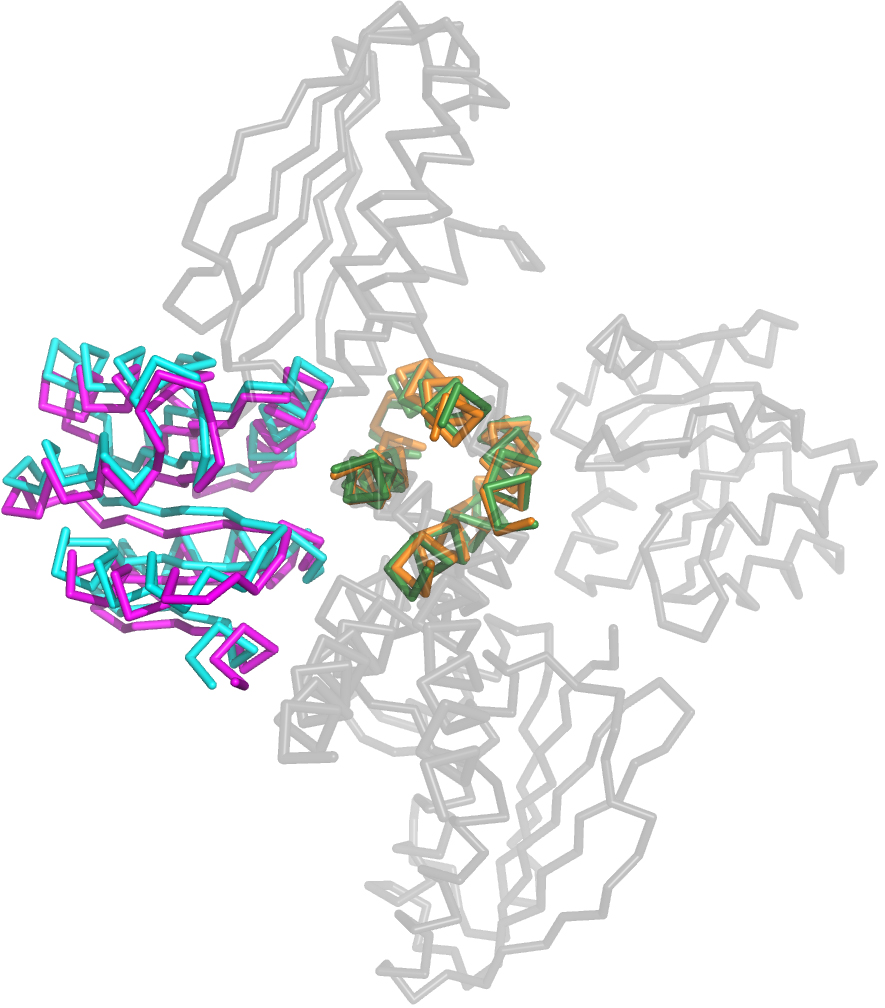
Figure 5. Structural superposition of the initial model of DesK:DesR used as Molecular Replacement probe and the final refined structure. Superposition of the Cα traces of the in silico-generated model of DesKDHp:DesRREC (orange:cyan) (Trajtenberg et al., 2014), onto the final refined crystal structure of DesKCSTAB:DesRREC (green:magenta). The entire proteins for the latter are observed in the crystal structure and could thus be manually built (indicated in grey). Of note, among the hundreds of in silico-generated models to be used for molecular replacement procedures, the few that proved useful in solving the structure, are close enough to the final structure as readily seen in this illustration, explaining their utility. Nonetheless, the actual experimental structure displays substantial changes, readily detectable in this view mostly along the DesRREC domains.
- Structure refinement is performed with Buster-TNT (Bricogne et al., 2009) using standard procedures, including non-crystallographic symmetry (NCS) restraints and Translation/Libration/Screw (TLS) descriptions in each model (documented in detail within the header of each atomic coordinates file available in the PDB).
Recipes
- LB medium
5 g/L NaCl
5 g/L yeast extract
15 g/L tryptone plus - 2x YT culture medium
5 g/L NaCl
10 g/L yeast extract
16 g/L tryptone plus - LB agar plates
300 ml LB medium
4.5 g agar - Lysis buffer
50 mM Tris-HCl pH 8
500 mM NaCl
EDTA-free cocktail of protease inhibitors - Immobilized Metal Affinity Chromatography (IMAC) binding and washing buffer
50 mM Tris-HCl pH 8
500 mM NaCl
40 mM imidazole
10% glycerol - IMAC elution buffer
50 mM Tris-HCl pH 8
500 mM NaCl
500 mM imidazole
10% glycerol - Dialysis buffer
50 mM Tris-HCl pH 8
300 mM NaCl
0.5 mM dithiothreitol - Size Exclusion Chromatography buffer for the phosphatase complex (SEC-P buffer)
20 mM Tris-HCl pH 8
500 mM NaCl
10 mM MgCl2 - Size Exclusion Chromatography buffer for the phosphotransferase complex (SEC-PT buffer)
20 mM Tris-HCl pH 8
300 mM NaCl
Acknowledgments
These protocols are adapted from previous work reported by our group (Trajtenberg et al., 2016). We are grateful to the staffs at synchrotron beamlines Proxima I and II, Soleil (France), especially William Shepard; and Daniela Albanesi for providing plasmid pHPKS/Pxyl-desKSTA. This work was supported by grants from Agencia Nacional de Investigación e Innovación (ANII), Uruguay (FCE2009_1_2679;FCE2007_219); Agence Nationale de la Recherche (ANR), France (PCV06_138918); Centro de Biología Estructural del Mercosur (www.cebem-lat.org) and Fondo para la Convergencia Estructural del MERCOSUR (COF 03/11). We are also grateful to the Institut Pasteur International Network for institutional support through the IMiZA International Joint Unit.
References
- Albanesi, D., Mansilla, M. C. and de Mendoza, D. (2004). The membrane fluidity sensor DesK of Bacillus subtilis controls the signal decay of its cognate response regulator. J Bacteriol 186(9): 2655-2663.
- Albanesi, D., Martin, M., Trajtenberg, F., Mansilla, M. C., Haouz, A., Alzari, P. M., de Mendoza, D. and Buschiazzo, A. (2009). Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc Natl Acad Sci U S A 106(38): 16185-16190.
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P., Sharff, A., Smart, O. S., Vonrhein, C. and Womack, T. O. (2009). BUSTER version 2.8.0. Global Phasing.
- Casino, P., Rubio, V. and Marina, A.(2009). Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139(2): 325-336.
- Chen, V. B., Arendall, W. B., 3rd, Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. and Richardson, D. C. (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66(Pt 1): 12-21.
- de Mendoza, D. (2014). Temperature sensing by membranes. Annu Rev Microbiol 68: 101-116.
- Emsley, P., Lohkamp, B., Scott, W. G. and Cowtan, K. (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66(Pt 4): 486-501.
- Evans, P. (2006). Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62(Pt 1): 72-82.
- Evans, P. R. (2011). An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr 67(Pt 4): 282-292.
- Gao, R. and Stock, A. M. (2009). Biological insights from structures of two-component proteins. Annu Rev Microbiol 63: 133-154.
- Kabsch, W. (2010). Xds. Acta Crystallogr D Biol Crystallogr 66(Pt 2): 125-132.
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. and Read, R. J. (2007). Phaser crystallographic software. J Appl Crystallogr 40(Pt 4): 658-674.
- Saita, E., Abriata, L. A., Tsai, Y. T., Trajtenberg, F., Lemmin, T., Buschiazzo, A., Dal Peraro, M., de Mendoza, D. and Albanesi, D. (2015). A coiled coil switch mediates cold sensing by the thermosensory protein DesK. Mol Microbiol 98(2): 258-271.
- Trajtenberg, F., Albanesi, D., Ruetalo, N., Botti, H., Mechaly, A. E., Nieves, M., Aguilar, P. S., Cybulski, L., Larrieux, N., de Mendoza, D. and Buschiazzo, A. (2014). Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation. MBio 5(6): e02105.
- Trajtenberg, F., Imelio, J. A., Machado, M. R., Larrieux, N., Marti, M. A., Obal, G., Mechaly, A. E. and Buschiazzo, A. (2016). Regulation of signaling directionality revealed by 3D snapshots of a kinase:regulator complex in action. Elife 5.
- Unger, T., Jacobovitch, Y., Dantes, A., Bernheim, R. and Peleg, Y. (2010). Applications of the Restriction Free (RF) cloning procedure for molecular manipulations and protein expression. J Struct Biol 172(1): 34-44.
- Vonrhein, C., Flensburg, C., Keller, P., Sharff, A., Smart, O., Paciorek, W., Womack, T. and Bricogne, G. (2011). Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr 67(Pt 4): 293-302.
Article Information
Copyright
Imelio et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Imelio, J. A., Larrieux, N., Mechaly, A. E., Trajtenberg, F. and Buschiazzo, A. A. (2017). Snapshots of the Signaling Complex DesK:DesR in Different Functional States Using Rational Mutagenesis and X-ray Crystallography. Bio-protocol 7(16): e2510. DOI: 10.21769/BioProtoc.2510.
- Trajtenberg, F., Imelio, J. A., Machado, M. R., Larrieux, N., Marti, M. A., Obal, G., Mechaly, A. E. and Buschiazzo, A. (2016). Regulation of signaling directionality revealed by 3D snapshots of a kinase:regulator complex in action. Elife 5.
Category
Microbiology > Microbial biochemistry > Protein > Structure
Biochemistry > Protein > Structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











