- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ribosome Fractionation in Yeast
Published: Vol 2, Iss 16, Aug 20, 2012 DOI: 10.21769/BioProtoc.251 Views: 22956

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Purification of Human Cytoplasmic Actins From Saccharomyces cerevisiae
Brian K. Haarer [...] Jessica L. Henty-Ridilla
Dec 5, 2023 1753 Views

In vitro Condensation Assay of Fluorescent Protein-Fused PRPP Amidotransferase Purified from Budding Yeast Cells
Masak Takaine
Jun 5, 2025 1786 Views

Fractionation and Extraction of Cell Wall Proteins From Candida albicans
Elizabeth Reyna-Beltrán [...] Juan Pedro Luna-Arias
Sep 20, 2025 2578 Views
Abstract
This protocol describes yeast ribosome fractionation in the gradient of sucrose. During the cyclic process of translation, a small (40S) and large (60S) ribosomal subunit associate with mRNA to form an 80S complex (monosome). This ribosome moves along the mRNA during translational elongation, and then dissociates into the 40S and 60S subunits on termination. During elongation by one ribosome, further ribosomes can initiate translation on the same mRNA to form polysomes. The mass of each polysomal complex is determined primarily by the number of ribosomes it contains. Hence, the population of polysomes within the cell can be size-fractionated by sucrose density gradient centrifugation on the basis of the loading of ribosomes on the mRNA. Several compounds help to maintain or to disrupt the polysomes (Figure 1).
Keywords: Ribosome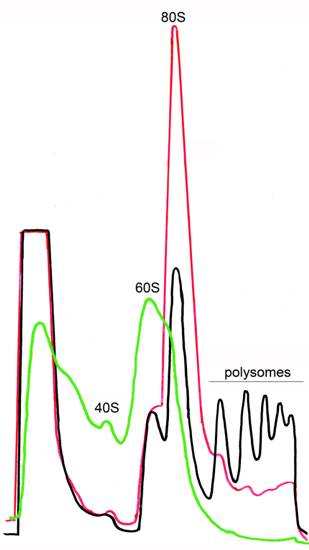
Figure 1. Ribosome profiles after sucrose gradient centrifugation. Ribosome fractionation was performed in the presence of CHX (black), in the absence of CHX (red) or in the presence of EDTA (green). CHX stabilizes polysomes. In the absence of CHX polysomes are destroyed but 80S is still present. 80S can be completely disrupted in cell extracts by treatment with EDTA.
Materials and Reagents
- Glass beads for cells breaking 0.5 mm (Bio Spec Products, catalog number: 110/9105 )
- Cycloheximide (CHX) (Sigma-Aldrich, catalog number: C7698 ) solution 100 mg/ml prepared on ethanol
- Bradford reactive (Bio-Rad Laboratories, catalog number: 500-0006 )
- Protease inhibitor cocktail (F. Hoffmann-La Roche, catalog number: 13560400 )
- Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7625 ) 100 mM solution prepared on isopropanol
- 100% trichloroacetic acid (TCA) (Fluka, catalog number: 91230 )
- Tubes for SW41 rotor Ti (Beckman Coulter, catalog number: 331372 )
- Protease inhibitor cocktail (F. Hoffmann-La Roche, catalog number: 1836170 )
- Lysis buffer
- Ethanol
- Laemmli sample buffer
- Bromophenol blue
- YPD media (see Recipes)
- Buffer A (see Recipes)
- 2x buffer B2 times concentrated (see Recipes)
- Sucrose gradient (see Recipes)
- Foni-inert (see Recipes)
Equipment
- Table Centrifuges
- Glass bead beater Genie Disruptor (Scientific Industries, catalog number: SI-DD38 )
- Ultracentrifuge
- Rotor SW41 Ti (Beckman Coulter, catalog number: 331362 )
- Density Gradient Fractionation System (ISCO, catalog number: 67-9000-177 )
Procedure
- Yeast cultures were grown on YPD or selective media. In the morning dilute night culture till OD600 = 0.15. Grow 100 ml of culture at 30 °C until OD600 = 0.6-0.8.
- To keep the polysomes add cycloheximide (CHX) till final concentration 0.1 mg/ml and incubate 10 min on ice.
- Spin the cells for 5 min at 4,000 x g and wash with 50 ml of cold water containing 0.1 mg/ml of CHX.
- Resuspend the pellets in 1 ml of lysis buffer containing 0.1 mg/ml of CHX, spin. Cells can be frozen and stored at -20 °C.
- Work on ice! Resuspend the pellet in 0.4 ml of buffer A, add 0.5 ml of glass beads and disrupt 15 min on the glass bead beater at 4 °C.
- To avoid the contamination with cells derby preclean the total extracts. For this transfer the liquid phase into the new tube. Centrifuge 1 min 16,000 x g at 4 °C.
- Transfer the supernatant into the new eppendorf tube and centrifuge 10 min 16,000 x g at 4 °C.
- Measure the total protein concentrations in the supernatants [= TE (Total Extracts)]. For this dilute the total extracts 1:20 and load 5 and 10 μl to the Bradford sample (0.8 ml of water and 0.2 ml of Bradford reactive).
Note: Lysate should be fresh, they can not be frozen! - Pour 12 ml gradients (7 to 47% sucrose) using Density Gradient Fractionation System (ISCO) (Figure 2). Gradients are best poured at the night before the centrifugation. Keep the gradients at 4 °C before loading samples. Balance gradient volumes in paired rotor buckets firstly by eye, carefully removing some of top of gradient using a pipette tip if necessary. Check with the balance.
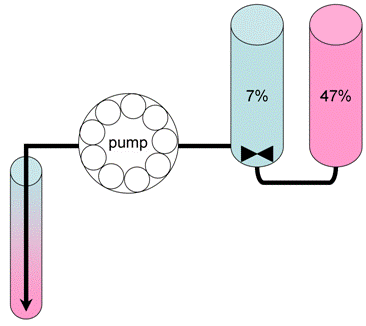
Figure 2. Preparation of sucrose density gradients for polysome analysis. 6 ml of 7% (marked in blue)and 6 ml of 47% (marked in pink) sucrose solutions are placed into the frontand back compartments of a gradient pourer, respectively. A small magnetic fleais used to mix the solution in the front compartment, which is drained slowly usinga peristaltic pump to the bottom of the centrifuge tube (approx 10 min per gradient).Care should be taken to minimize the mixing in the centrifuge tube. - Load 0.2 ml of the total extracts containing 2-4 mg of total protein on the top of 7-47% sucrose gradient without disturbing the surface. Note: Do not load more than 0.25 ml of the total extracts!
- Centrifuge for 150 min at 220,000 x g (39,000 rpm for rotor SW41) at 4 °C.
- Before spin is complete, set up to analyze gradient(s). Standardize absorbance detector with a 1x buffer B. Set baseline according to the Instruction Manual.
Note: Before putting first gradient on make sure the pump line is filled with foni-inert so that no bubbles can enter sample - this will destroy the gradient! Try briefly raising the needle and running 1 ml of foni-inert into the base cup just prior to first fractionation. - Carefully remove gradients and store at 4 °C without disturbing. Gradients must be fractionated before they start to diffuse. Insert first gradient snugly into top of fractionator [Density Gradient Fractionation System (ISCO)] and raise lower platform to base of tube, and screw-lock. Slowly screw up needle into bottom of gradient tube (Figure 3). Run the pump.
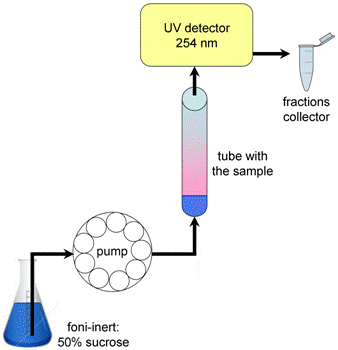
Figure 3. Fractionation of polysome gradients. Pierce the tube with theneedle. Using a peristaltic pump, extrude the gradient from the tube with afoni-inert through a spectrophotometer that monitors the absorbance at 254 nm,and then fractionated into eppendorf tubes. - End run when first droplets of foni-inert are collected at end of gradient, then put the pump in the reverse mode and pump back most of the foni-inert for reuse. Be careful that no new air bubbles are introduced into pump line and needle during this procedure.
- Clean up after fractionating all parts of the gradient fractionation system. Rinse entire system with water, then with 20% ethanol. Clean up bench and equipment. Discard waste solutions and trash.
Density Gradient Fractionation System (ISCO)
Isco Type 6 Optical Unit:
Use 254 nm filter and source screen
UA-5 Absorbance Detector Settings:
Chart Speed: 30-60 cm/h
Sensitivity: 1.0 or 2.0 (A260 Units = Full scale)
Slope Sensitivity = High
Pump: Setting 40, clockwise
Fraction Collector: (if desired)
Use 1.5 ml microfuge tubes inside larger glass tubes
Collect 0.4 min drops (0.75 ml) - Take 0.4 ml of fraction (the rest freeze) and add 60 μl of 100% TCA, incubate for 10 min on ice.
- Spin at 16,000 x g for 15 min at 4 °C.
- Aspirate all traces of TCA and resuspend the pellets in 50 μl of Laemmli sample buffer.
- Load 10 μl on SDS-PAGE
Note: In case of polysomes disruption, polysomes were dissociated by treatment with 25 mM EDTA added instead of CHX, in the buffer A and in the gradient.
Recipes
- YPD media
2% glucose
2% bactopeptone
1% yeast extract - Buffer A
20 mM Hepes (pH 8.0)
50 mM KCl
10 mM MgCl2
1 % Triton X-100
1 mM DTT
1 mM PMSF
CHX 0.1 μg/ml
Protease inhibitor cocktail 1 tablet for 10 ml of extract - 2x buffer B2 times concentrated
40 mM Hepes
100 mM KCl
20 mM MgCl2 - Sucrose gradient
7% sucrose on buffer B (1x) + CHX 0.1 mg/ml
47% sucrose on buffer B (1x) + CHX 0.1 mg/ml - Foni-inert
50% sucrose on water
0.01% bromophenol blue
Acknowledgments
This work was supported by grant from Ernst and Lucie Schmidheiny Foundation and Pierre Mercier Foundation awarded to O.O.P. and grants 31003A-120419 and 31003A_135794 of the Swiss National Science Foundation as well as a grant from the Novartis Foundation awarded to M.A.C.
References
- Albanese, V., Reissmann, S. and Frydman, J. (2010). A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J Cell Biol 189(1): 69-81.
- Panasenko, O. O. and Collart, M. A. (2012). Presence of Not5 and ubiquitinated Rps7A in polysome fractions depends upon the Not4 E3 ligase. Mol Microbiol 83(3): 640-653.
Article Information
Copyright
© 2012 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Panasenko, O. O. (2012). Ribosome Fractionation in Yeast. Bio-protocol 2(16): e251. DOI: 10.21769/BioProtoc.251.
Category
Microbiology > Microbial cell biology > Organelle isolation
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










