- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection of Pathogens and Ampicillin-resistance Genes Using Multiplex Padlock Probes
Published: Vol 7, Iss 16, Aug 20, 2017 DOI: 10.21769/BioProtoc.2504 Views: 10268
Reviewed by: Modesto Redrejo-RodriguezAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Protocol to Retrieve Unknown Flanking DNA Using Fork PCR for Genome Walking
Hongjing Wu [...] Haixing Li
Jan 20, 2025 2407 Views

Protocol to Identify Unknown Flanking DNA Using Partially Overlapping Primer-based PCR for Genome Walking
Mengya Jia [...] Haixing Li
Feb 5, 2025 1380 Views

Protocol to Mine Unknown Flanking DNA Using PER-PCR for Genome Walking
Zhou Yu [...] Haixing Li
Feb 20, 2025 1509 Views
Abstract
Diagnostic assays for pathogen identification and characterization are limited either by the number of simultaneously detectable targets, which rely on multiplexing methods, or by time constraints due to cultivation-based techniques. We recently presented a 100-plex method for human pathogen characterization to identify 75 bacterial and fungal species as well as 33 clinically relevant β-lactamases (Barišić et al., 2016). By using 16S rRNA gene sequences as barcode elements in the padlock probes, and two different fluorescence channels for species and antibiotic resistance identification, we managed to cut the number of microarray probes needed by half. Consequently, we present here the protocol of an assay with a runtime of approx. 8 h and a detection limit of 105 cfu ml-1. A total of 89% of β-lactamases and 93.7% of species were identified correctly.
Keywords: Multiplex detectionBackground
β-Lactamases are a class of antibiotic resistance genes which provide resistance to β-lactam antibiotics, which structurally mimic D-alanyl-D-alanine, a component of the bacterial cell wall and thereby inhibit bacterial cell wall synthesis. β-Lactamases are able to hydrolyze the central component of β-lactam antibiotics, the β-lactam ring, and render them useless (Kong et al., 2010). Today, over 1,000 β-lactamases are described and a huge potential environmental reservoir exists (Bush, 2010; Brandt et al., 2017). β-Lactamases are ancient enzymes and we classify them as class A, C, and D (serine β-lactamases) with a serine catalytic site, or as class B (metallo-β-lactamases) whose active center is zinc-dependent (Hall and Barlow, 2003 and 2004). Despite their high phylogenetic age, the serine β-lactamases probably share a common ancestor and acquired a high number of SNPs due to a permanent selection pressure. Additionally, to β-lactamases, more than 500 other antibiotic resistance genes exist (Zankari et al., 2012).
Current multiplexing methods reduce the number of simultaneously detectable targets while cultivation-dependent techniques are limited by time constraints. Given these facts and the high number of pathogens of clinical importance, new methods are needed for the fast characterization and identification of pathogens, virulence factors and antibiotic resistance genes.
The gold standard of infection diagnostics takes 2-3 days and is cultivation-dependent (Marik, 2014). Additionally, PCR methods provide results at a high sensitivity and low cost, but remain impractical due to the high number of clinically relevant targets (Mussap et al., 2007; Wellinghausen et al., 2009). The current multiplex-PCR protocols are not suitable for a high number of targets and the limitations can only be overcome by microfluidic-based assays, which run a high number of analyses in parallel.
Padlock probes are linear DNA probes, which upon annealing, circularize and are then used for rolling circle amplifications (Nilsson et al., 1994; Hardenbol et al., 2005). They allow for a higher number of multiplexing and can easily be integrated into established PCR-based assays. Recently, we presented a 100-plex method based on padlock probes for pathogen characterization (75 bacterial and fungal species) as well as 33 clinically important β-lactamases (Barišić et al., 2016). By adapting this method from our previous work (Barišić et al., 2013), we increased the sensitivity and specificity of the assay and we managed to cut down the number of microarray probes needed by half by using 16S rRNA sequences as barcode elements in the padlock probes and two different fluorescence channels for species identification and antibiotic resistance characterization. Here, we present an assay to overcome time limitations and to increase the number of detectable targets. Our assay allows for the detection of up to 105 cfu ml-1 in a total of 8 h. We were able to retrieve and correctly characterize 89% of β-lactamases and to identify 93.7% of all species.
Materials and Reagents
- Filter tips 10 µl, 20 µl, 200 µl and 1,250 µl (e.g., Biozym, catalog numbers: VT0200 , VT0220 , VT0240 and VT0270 )
- Safe-Lock tubes 1.5 ml (Eppendorf, catalog number: 022363204 )
- Falcon 15 ml conical centrifuge tubes (Corning, catalog number: 352196 )
- 2 ml screw-cap tube (e.g., Roche Molecular Systems, catalog number: 03358941001 )
- Ritter Riplate 384 well plate PP (Ritter, catalog number: 43001-0035 )
- Glass beads, acid-washed, 150-212 μm (Sigma-Aldrich, catalog number: G1145 )
- Glass beads, acid-washed, 425-600 μm (Sigma-Aldrich, catalog number: G8772 )
- Vantage Silylated Aldehyde Slides (CEL Associates, catalog number: VSS-25 )
- LifterSlip mSeries cover slips for microarray slides, 55 µl (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 25X60IM5439001LS )
- PCR tubes 0.2 ml (Eppendorf, catalog number: 0030124537 )
- Millex-GV 0.22 µm syringe filter units (Merck, catalog number: SLGV033RS )
- 50 ml tubes (e.g., Corning, catalog number: 430829 )
- Disposable syringes, e.g., Omnifix 50 ml LL (B. Braun Medical, catalog number: 8508577FN )
- The multiplex PCR primers (66 in total, Table S1) targeting the β-lactamase genes were ordered from Microsynth (Balgach, Switzerland) (Note 3)
- The padlock probes (66 in total, Figure 1, Table S2) targeting the β-lactamases were ordered from Integrated DNA Technologies (Coralville, IA, USA)
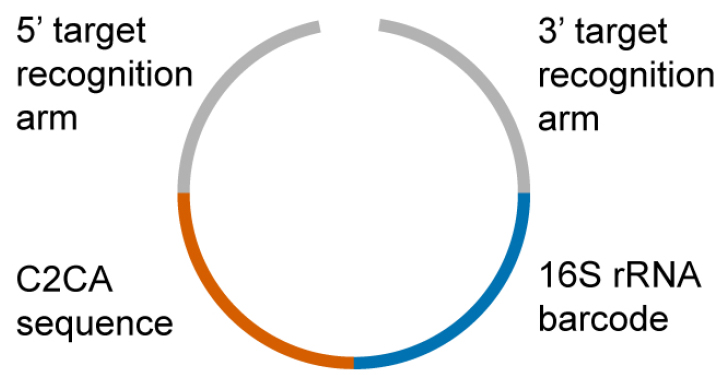
Figure 1. Schematic illustration of a padlock probe. The 3’ and the 5’ target recognition arms bind to the multiplex PCR products of the β-lactamase genes. During the binding process, the padlock probes are circularized and subsequently ligated. The maximum distance of the padlock probe binding region to the 3’ or 5’-ends of the PCR product should not exceed 200 base pairs. Since the padlock probes gets concatenated with the PCR product upon the ligation reaction, longer distances cause an inhibition of subsequent RCAs. The C2CA sequence is needed for the circle-to-circle amplification and comprises an AluI restriction site for monomerization of the amplification products. The barcode sequence is derived from the 16S rRNA gene. This allowed us to halve the number of microarray probes because the C2CA products and 16S rRNA gene PCR products are detected on the same microarray probe but in different fluorescence channels. - The 5’-amino-modified microarray probes (274 in total, Table S3) were ordered from Microsynth (Balgach, Switzerland) (Note 3)
- Nuclease-free water for PCR application comes with the Mastermix 16S Basic kit or can be purchased separately (e.g., Fresenius Kabi, Aqua bidest. ‘Fresenius’, no catalog number)
- Ultrapure water, hereafter simply referred to as water or H2O (Note 1)
- The universal bacterial 16S rRNA primers 45f++ (5’-GCYTAAYACATGCAAGTCGARCG-3’) and 783R (5’-TGGACTACCAGGGTATCTAATCCT-3’) were ordered from Integrated DNA Technologies (Coralville, IA, USA)
- The fungal 18S rRNA (ITS region) primers ITS3 (5’-GCATCGATGAAGAACGCAGC-3’) and ITS4+ (5’-TCCT-CCGCTTATTGATATGCTTAAGT-3’) were ordered from Integrated DNA Technologies (Coralville, IA, USA)
- The circle-to-circle amplification (C2CA) oligonucleotides C2CA- (5’-TACTCGAGGAGCTGCATACAC-3’) and C2CA+ (5’-GTGTATGCAGCTCCTCGAGTA-3’) were ordered from Integrated DNA Technologies (Coralville, IA, USA)
- The Cy5-labelled hybridization control (complimentary to ‘Bsrev’) (5’-Cy5-AAGCTCACTGGCCGTCGTTTTAAA-3’) was ordered from Microsynth (Balgach, Switzerland)
- T4 DNA ligase (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: EL0011 )
- T4 polynucleotide kinase (10 U/µl) supplied with 10x reaction buffer A (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: EK0031 )
- ATP solution (100 mM) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0441 )
- Mastermix 16S Basic, DNA-free (Molzym, catalog number: S-040-0250 ) containing a 2.5x complete master mix, Moltaq 16S DNA polymerase and PCR-grade water (Note 2)
- dNTP mix (10 mM each) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0192 )
- Cy5-dCTP (1 mM solution) (GE Healthcare, catalog number: PA55021 )
- VentR (exo-) DNA polymerase (New England Biolabs, catalog number: M0257S ) supplied with 10x ThermoPol reaction buffer and 100 mM MgSO4
- ExpressHyb hybridization solution (Takara Bio, Clontech, catalog number: 636831 )
- Ampligase thermostable DNA ligase (5 U/µl) and Ampligase 10x reaction buffer (Epicentre, catalog number: A32750 )
- Bovine serum albumin (BSA), molecular biology grade (New England Biolabs, catalog number: B9000S )
- phi29 DNA polymerase supplied with 10x phi29 DNA polymerase reaction buffer (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: EP0091 )
- AluI restriction enzyme (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: ER0011 )
- Atto532-dCTP (MoBiTec, Göttingen, Germany)
- SDS solution 10% for molecular biology (AppliChem, catalog number: A0676 )
- Tryptic soy broth, also referred to as CASO medium (Casein-peptone soymeal-peptone broth) (Merck, catalog number: 105459 )
- PBS (10x), pH 7.2 (Thermo Fisher Scientific, GibcoTM, catalog number: 70013 )
- Betaine monohydrate (Sigma-Aldrich, catalog number: B2754 )
- UltraPure SSC, 20x (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15557036 )
- CASO medium (see Recipes)
- 1x phosphate-buffered saline (PBS) (see Recipes)
- 2x spotting buffer (6x SSC, 3 M betaine) (see Recipes)
Equipment
- Pipettes (e.g., Sartorius, catalog numbers: 728020 , 728050 , 728060 and 728070 )
- Microbiological incubator shaker (e.g., IKA, model: KS 4000 i control )
- Tabletop centrifuge for 1.5 ml tubes (e.g., Eppendorf, model: 5424 )
- Roche MagNA Lyser Instrument (Basel, Switzerland)
- Thermomixer comfort (Eppendorf, Hamburg, Germany)
- Epoch Microplate spectrophotometer (Biotek, Winooski, VT, USA)
- Biosan DNA/RNA UV-cleaner box (Warren, MI, USA) (recommended, see Note 4)
- Thermal cycler (e.g., Thermo Fisher Scientific, Applied Biosystems, model: GeneAmpTM PCR System 2700 ) (Paisley, UK)
- Heraeus Megafuge 40R with a TX-750 rotor (Thermo Fisher Scientific, Thermo ScientificTM, model: HeraeusTM MegafugeTM 40R , catalog number: 75004518; TX-750 rotor: Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 75003180 ) and inserts for plates (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 75003617 ) and Falcon tubes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 75003608 )
- Slide humidity incubation box (e.g., LabScientific, catalog number: HIC-3 )
- GeneMachines Omnigrid 100 contact arrayer (Madison, WI, USA)
- International Microarray Pin Stealth 3 SMP3 (Telechem, catalog number: SMP3 )
- Agilent SureScan DNA Microarray Scanner (Santa Clara, CA, USA)
- Autoclave
- Sartorius arium pro UV ultrapure water system (Sartorius, model: arium® pro )
Software
- GenePix Pro 6.0 Software (Molecular Devices LLC, Sunnyvale, CA, USA)
- Microsoft Excel or any other data analysis software
- ARB software package for microarray probe design (Note 3)
- Primer3 for primer design (Note 3)
Procedure
- Preparation of oligonucleotides
- Dissolve the 5’ amino modified microarray probes to 100 µM in nuclease-free water.
- Dissolve the 33-plex PCR primers to 100 µM in nuclease-free water, then create a 100 µM primer mix by pipetting equal volume of all 66 primers (e.g., 6 µl each) into one 1.5 ml Eppendorf tube.
- Dissolving and phosphorylation of the padlock probes (Note 8).
- Dissolve the padlock probes to 200 µM in nuclease-free water, then create a 2 µM per probe Master mix by pipetting 1 volume of padlock probe (e.g., 66 x 10.00 µl) and 0.34 volumes of nuclease-free water (e.g., 66 x 10.00 x 0.515 = 340.00 µl in total) into a 1.5 ml Eppendorf tube.
- Phosphorylation of the padlock probe Master mix is done by combining 10 µl of T4 polynucleotide kinase (0.2 U/µl final conc.), 50 µl of 10x reaction buffer A (1x final conc.), 50 µl of 10 mM ATP (1 mM final conc.), 250 µl of the padlock probe mix (1 µM per probe final conc.) and 140 µl PCR-grade water (final volume 500 µl).
- Incubate for 30 min at 37 °C, then for 10 min at 65 °C.
- Dissolve the padlock probes to 200 µM in nuclease-free water, then create a 2 µM per probe Master mix by pipetting 1 volume of padlock probe (e.g., 66 x 10.00 µl) and 0.34 volumes of nuclease-free water (e.g., 66 x 10.00 x 0.515 = 340.00 µl in total) into a 1.5 ml Eppendorf tube.
- Store microarray probes, padlock probes, primers and the respective master mixes at -20 °C.
- Dissolve the 5’ amino modified microarray probes to 100 µM in nuclease-free water.
- Growth of bacterial cells and preparation of genomic DNA extracts (Note 5)
- Inoculate a pure culture (e.g., from a glycerol stock) or a clinical isolate (e.g., from an agar culture) in approx. 5 ml CASO medium (see Recipes) in a 15 ml Falcon tube (Note 6). Do not completely screw the cap to allow for proper aeration of the bacteria.
- Incubate samples for 14-18 h at 37 °C in an orbital shaking incubator at 300 rpm (Note 7).
- Vortex the Falcon tube and transfer 2 ml of the culture to a 2 ml Eppendorf tube.
- Pellet the bacteria by centrifugation for 5 min at 2,400 x g at room temperature.
- Resuspend the pellet in 1 ml 1x PBS (see Recipes).
- Centrifuge the suspension for 5 min at 2,400 x g at room temperature.
- Resuspend the pellet in 1 ml sterile ddH2O and transfer to a 2 ml screw-cap tube suitable for the MagNA Lyser.
- Add acid-washed beads of 150-212 µm and acid-washed beads of 425-600 µm diameter at equal volumes to the tube, so that the beads make up about one third of the tube.
- Disrupt the bacterial and fungal cells using a MagNA Lyser for 30 sec at 6,500 rpm at room temperature.
- Leave sample 5 min at room temperature.
- Repeat step A9 once.
- For thermal lysis, incubate the sample for 20 min at 95 °C in a thermomixer.
- Centrifuge for 10 min at maximum speed.
- Transfer the supernatant to a new tube.
- Inoculate a pure culture (e.g., from a glycerol stock) or a clinical isolate (e.g., from an agar culture) in approx. 5 ml CASO medium (see Recipes) in a 15 ml Falcon tube (Note 6). Do not completely screw the cap to allow for proper aeration of the bacteria.
- Microarray processing
- Prepare the Ritter 384-well spotting plate by adding 18 µl of the 2x spotting buffer (see Recipes) to every well to be used, and then add 18 µl of microarray oligonucleotide to every well (final concentration of every oligonucleotide is now 50 µM in 1x spotting buffer). Take note of which oligonucleotides are in which wells and how your spotter transfers the oligonucleotides to the glass slides (Note 4).
- Set the Omnigrid contact arrayer to an air humidity of 60% and set up the machine to print the oligonucleotides in 4 replicates per slide (Notes 9 and 10).
- Spot the oligonucleotides onto glass slides with aldehyde surface using SMP3 pins which produce spots with 100 µm in diameter.
- After spotting, allow the oligonucleotides to attach for approx. 5 h in the humid atmosphere.
- The glass slides are not washed after spotting.
- Store the slides at room temperature.
- Prepare the Ritter 384-well spotting plate by adding 18 µl of the 2x spotting buffer (see Recipes) to every well to be used, and then add 18 µl of microarray oligonucleotide to every well (final concentration of every oligonucleotide is now 50 µM in 1x spotting buffer). Take note of which oligonucleotides are in which wells and how your spotter transfers the oligonucleotides to the glass slides (Note 4).
- Species identification
- 16S rRNA gene PCR
- Prepare the PCR master mix in 0.2 ml PCR tubes for n + 1 reactions, using the Molzym Mastermix 16S Basic kit, according to Table 1. Include at least one negative control (NTC) where you replace the genomic DNA extract with water. A primer mix consisting of equal quantities of all four primers at 15 µM can be prepared in advance to avoid repetitive pipetting. In this case, 1.20 µl of the primer mix is used. Keep samples chilled on ice until they are placed into the thermal cycler (Note 11).
Table 1. Multiplex PCR Master mix for 16S rRNA gene identification
- Run this reaction in parallel with ‘33-plex pre-amplification PCR for β-lactamase-encoding gene identification’ using the PCR program outlined in Table 2 in a thermal cycler with heated lid (Figure 2).
- Keep the samples at 4 °C after the PCR run is finished.
Table 2. Multiplex PCR program for 16S rRNA gene identification and for pre-amplification of β-lactamase-encoding gene information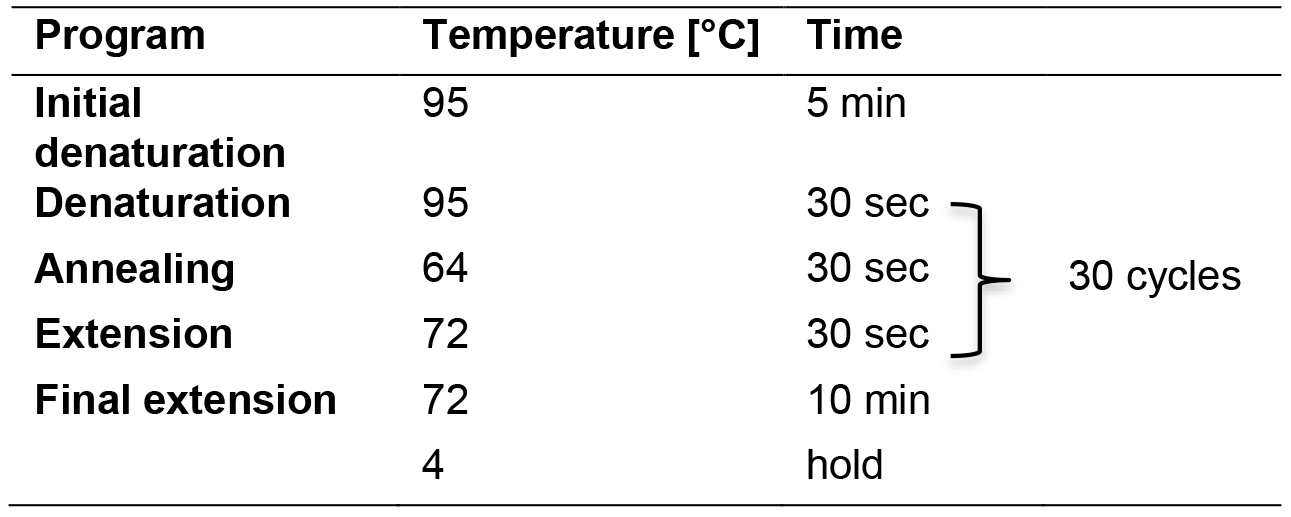
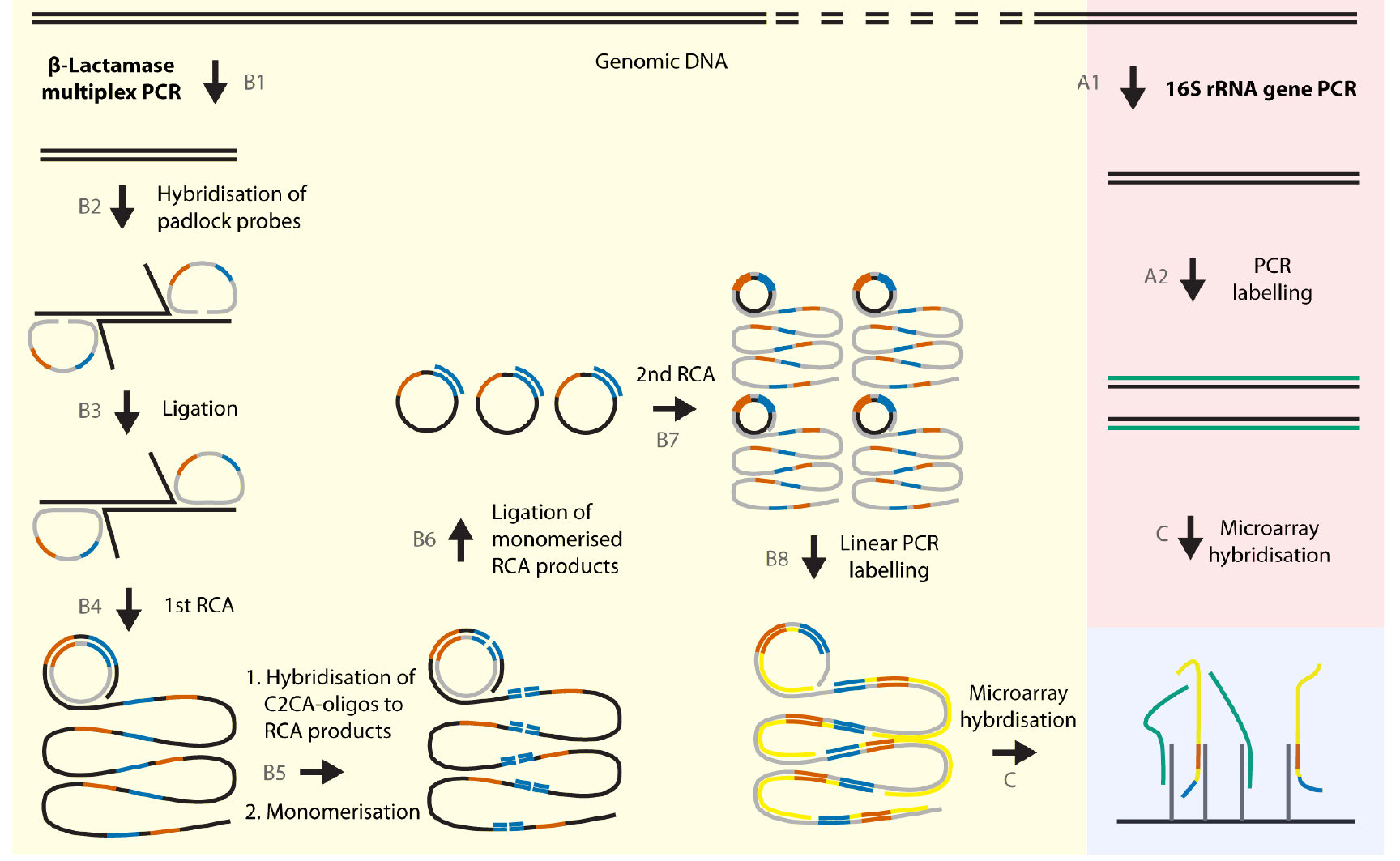
Figure 2. Schematic illustration of the detection principle of the assay. For the species identification, the 16S rRNA gene is amplified using universal 16S primers (A1). Subsequently, the PCR products are labeled using a linear PCR and Cy5-modified cytosines (A2). In parallel, β-lactamase genes are pre-amplified in a 33-plex PCR (B1). Then, the padlock probes are hybridized (B2) and ligated (B3) using the β-lactamase PCR products as template and amplified in a first rolling circle amplification (RCA) (B4). The RCA products comprise a C2CA sequence and corresponding 16S rRNA barcode sequences. In a next reaction, the RCA products are monomerized (B5) using a restriction enzyme and circularized (B6) to serve as a new template for a second RCA (B7). These C2CA products are labelled in a linear PCR reaction using Atto532-modified cytosines (B8). Finally, the labelled amplification products from the 16S rRNA gene PCR and from the multiplex padlock assay are pooled together and hybridized to a microarray detecting the products at two different fluorescence wavelengths (C).
- Prepare the PCR master mix in 0.2 ml PCR tubes for n + 1 reactions, using the Molzym Mastermix 16S Basic kit, according to Table 1. Include at least one negative control (NTC) where you replace the genomic DNA extract with water. A primer mix consisting of equal quantities of all four primers at 15 µM can be prepared in advance to avoid repetitive pipetting. In this case, 1.20 µl of the primer mix is used. Keep samples chilled on ice until they are placed into the thermal cycler (Note 11).
- Linear PCR for labelling of the PCR products
- Prepare the linear PCR master mix in 0.2 ml PCR tubes for n + 1 reactions according to Table 3 with the PCR products from the previous PCR. Keep samples chilled until placed in the thermal cycler.
Table 3. Linear PCR Master mix for labelling of the PCR products
- Run this reaction in parallel with ‘Labelling of C2CA products in a linear PCR’ using the PCR program outlined in Table 4 in a thermal cycler with heated lid.
Table 4. Linear PCR program for labelling of the PCR and C2CA products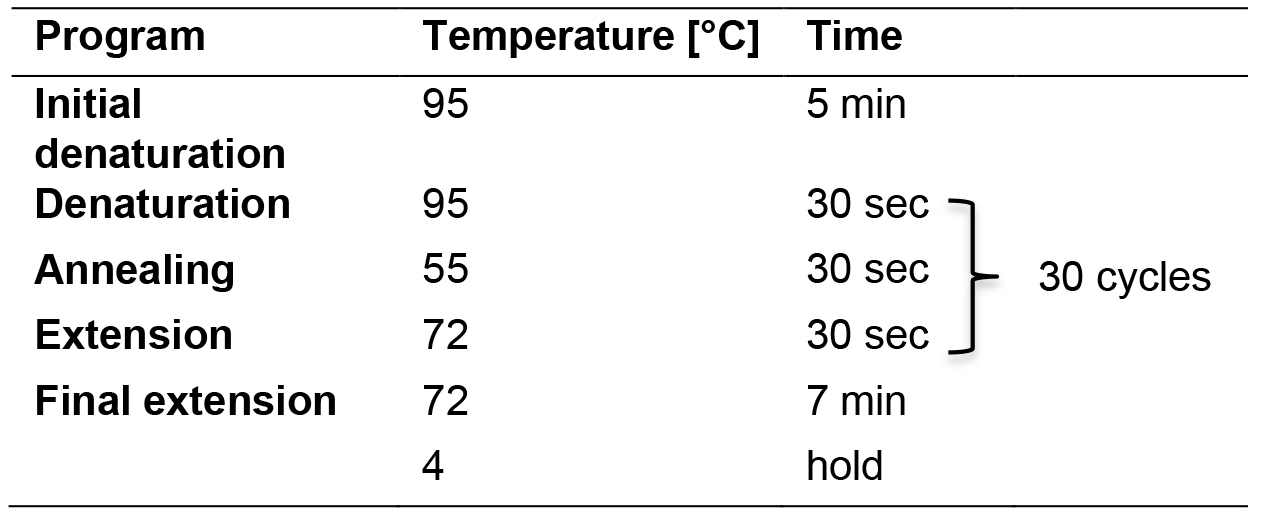
- Prepare the linear PCR master mix in 0.2 ml PCR tubes for n + 1 reactions according to Table 3 with the PCR products from the previous PCR. Keep samples chilled until placed in the thermal cycler.
- 16S rRNA gene PCR
- Multiplex β-lactamase-encoding gene identification
- 33-plex pre-amplification PCR for β-lactamase-encoding gene identification
- Prepare the PCR master mix in 0.2 ml PCR tubes for n + 1 reactions, using the Molzym Mastermix 16S Basic kit, according to Table 5. Include at least one NTC. The primer concentration corresponds to 76 nM each. Keep samples chilled until placed in the thermal cycler (Note 11).
Table 5. Multiplex pre-amplification PCR for β-lactamase-encoding gene identification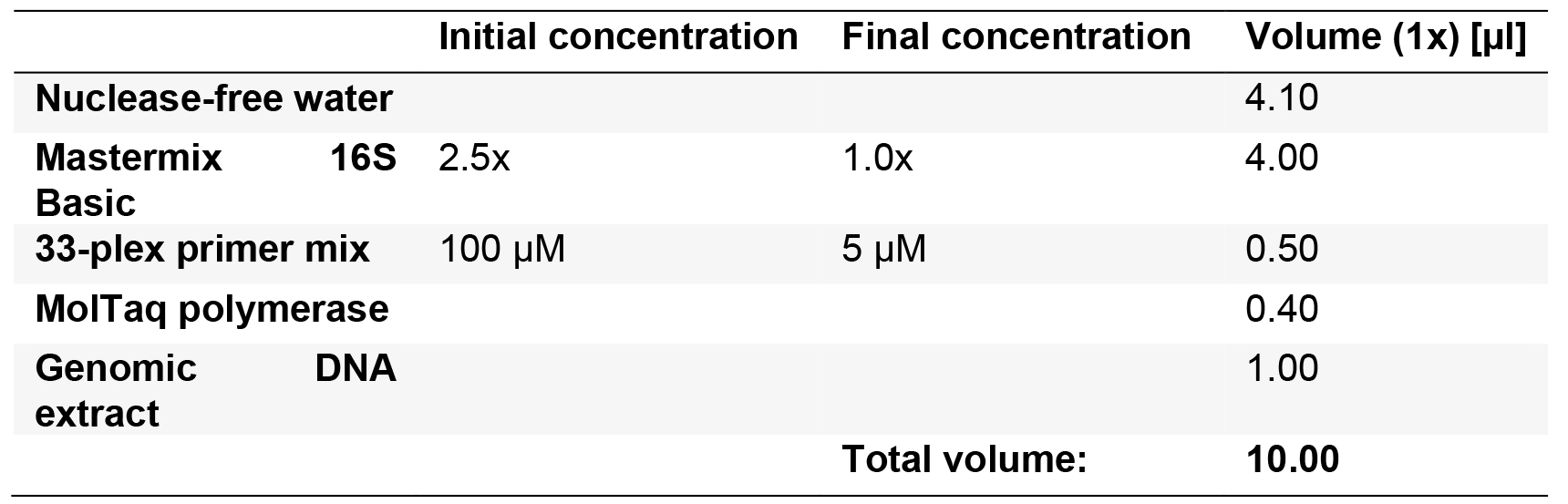
- Run the PCR program as described in Table 2.
- Prepare the PCR master mix in 0.2 ml PCR tubes for n + 1 reactions, using the Molzym Mastermix 16S Basic kit, according to Table 5. Include at least one NTC. The primer concentration corresponds to 76 nM each. Keep samples chilled until placed in the thermal cycler (Note 11).
- Hybridization and circularization by ligation of padlock probes
- Prepare the ligation mixture (10 µl per sample) according to Table 6 for n + 1 samples.
Table 6. Enzymatic reaction mix for circularization by enzymatic ligation of padlock probes. A padlock master mix concentration of 66 nM equals to 1 nM per probe.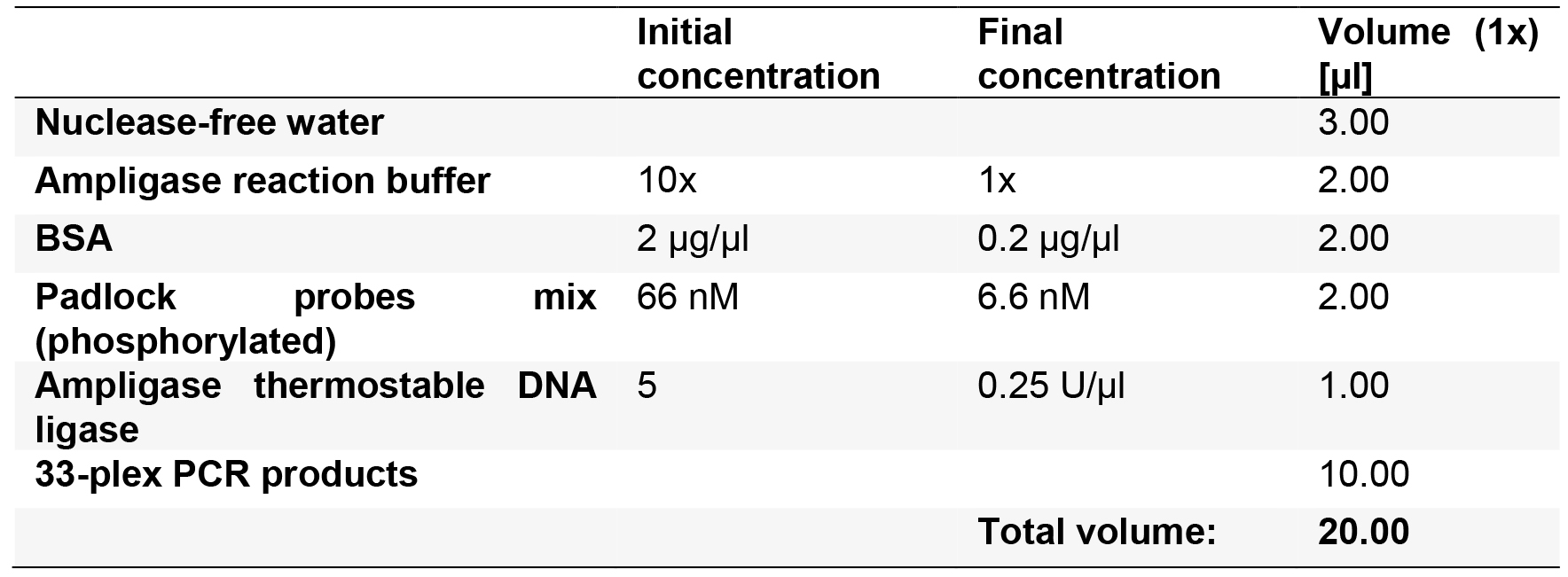
- Add 10 µl of the ligation mixture to the PCR products from the 33-plex PCR. The final concentration of every padlock probe is 100 pM each.
- Incubate at 95 °C for 5 min, then at 60 °C for 1 h in a thermal cycler with heated lid.
- Prepare the ligation mixture (10 µl per sample) according to Table 6 for n + 1 samples.
- First rolling circle amplification
- Prepare the reaction Master mix for the first rolling circle amplification (RCA) as described in Table 7, using the ligation products from the previous reaction, for n + 1 reactions.
Table 7. Reaction mix for the first RCA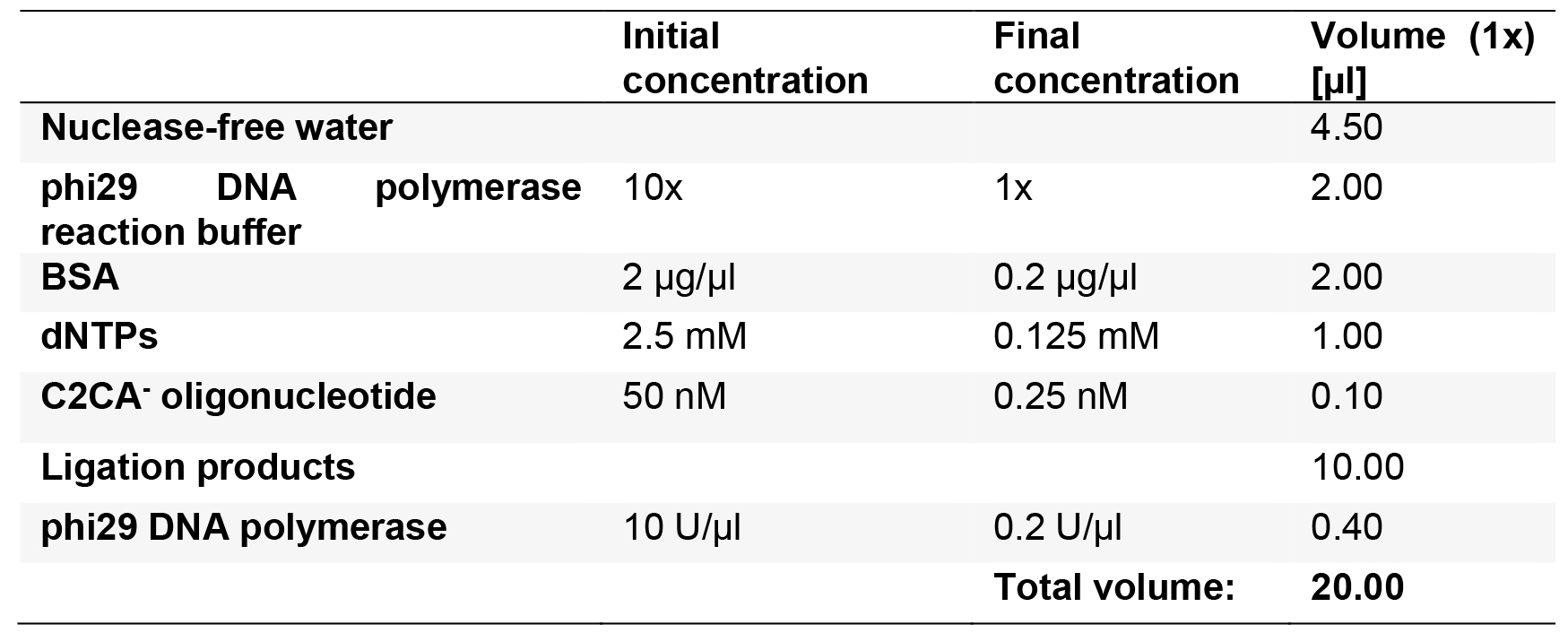
- Incubate for 20 min at 37 °C.
- Inactivate the polymerase by a 2 min incubation at 65 °C.
- Prepare the reaction Master mix for the first rolling circle amplification (RCA) as described in Table 7, using the ligation products from the previous reaction, for n + 1 reactions.
- Hybridization of C2CA oligonucleotides and monomerization by enzymatic restriction
- Prepare the restriction mixture as described in Table 8 for n + 1 reactions.
Table 8. Reaction mix for the enzymatic restriction reaction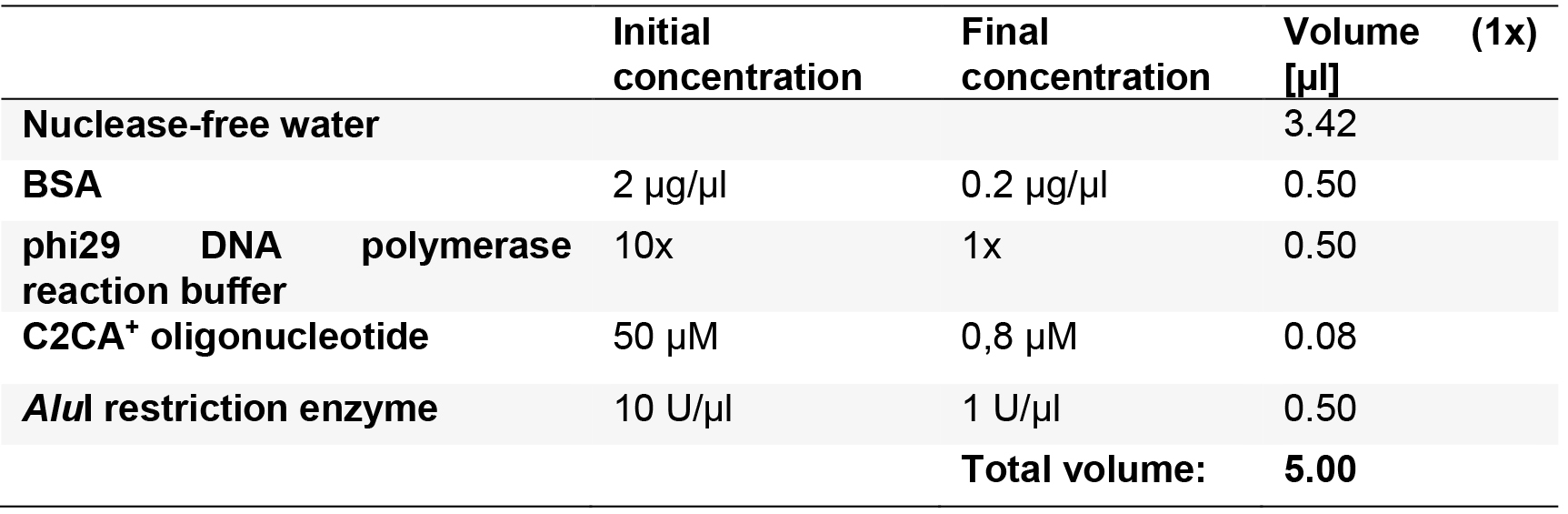
- Add 5 µl of the restriction mixture to the first RCA mixture (total volume is now 25 µl).
- Incubate at 37 °C for 5 min.
- Incubate at 65 °C for 5 min to inactivate the restriction enzyme.
- Prepare the restriction mixture as described in Table 8 for n + 1 reactions.
- Circularization, ligation and second RCA
- Prepare the ligation and second RCA Master mix as described in Table 9 for n + 1 reactions using the products from the previous reaction.
Table 9. Circularization, ligation and second RCA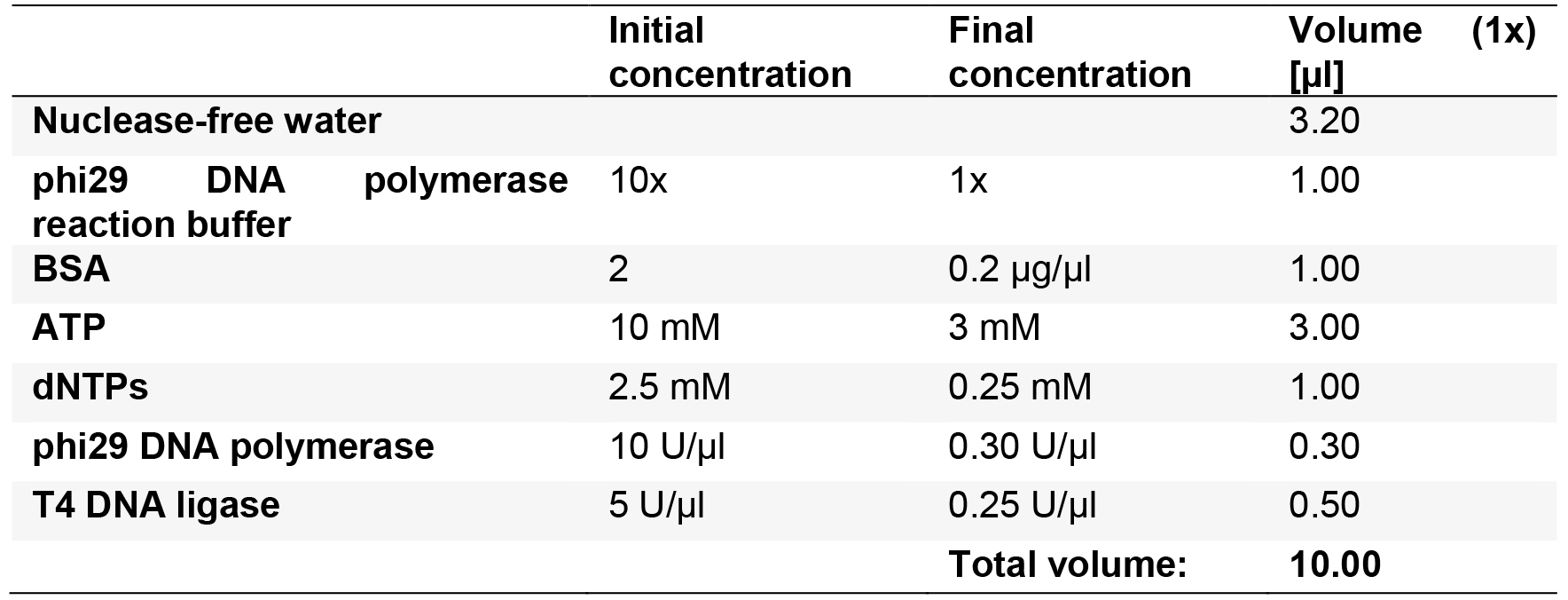
- Add 10 µl of the master mix to the previous reaction.
- Incubate for 20 min at 37 °C.
- Inactivate the ligation and RCA reaction by incubation for 2 min at 65 °C.
- Prepare the ligation and second RCA Master mix as described in Table 9 for n + 1 reactions using the products from the previous reaction.
- Labelling of C2CA products in a linear PCR
- Prepare the linear PCR master mix as described in Table 10 for n + 1 reactions and finally add 6 µl C2CA products.
Table 10. Labelling of C2CA products in a linear PCR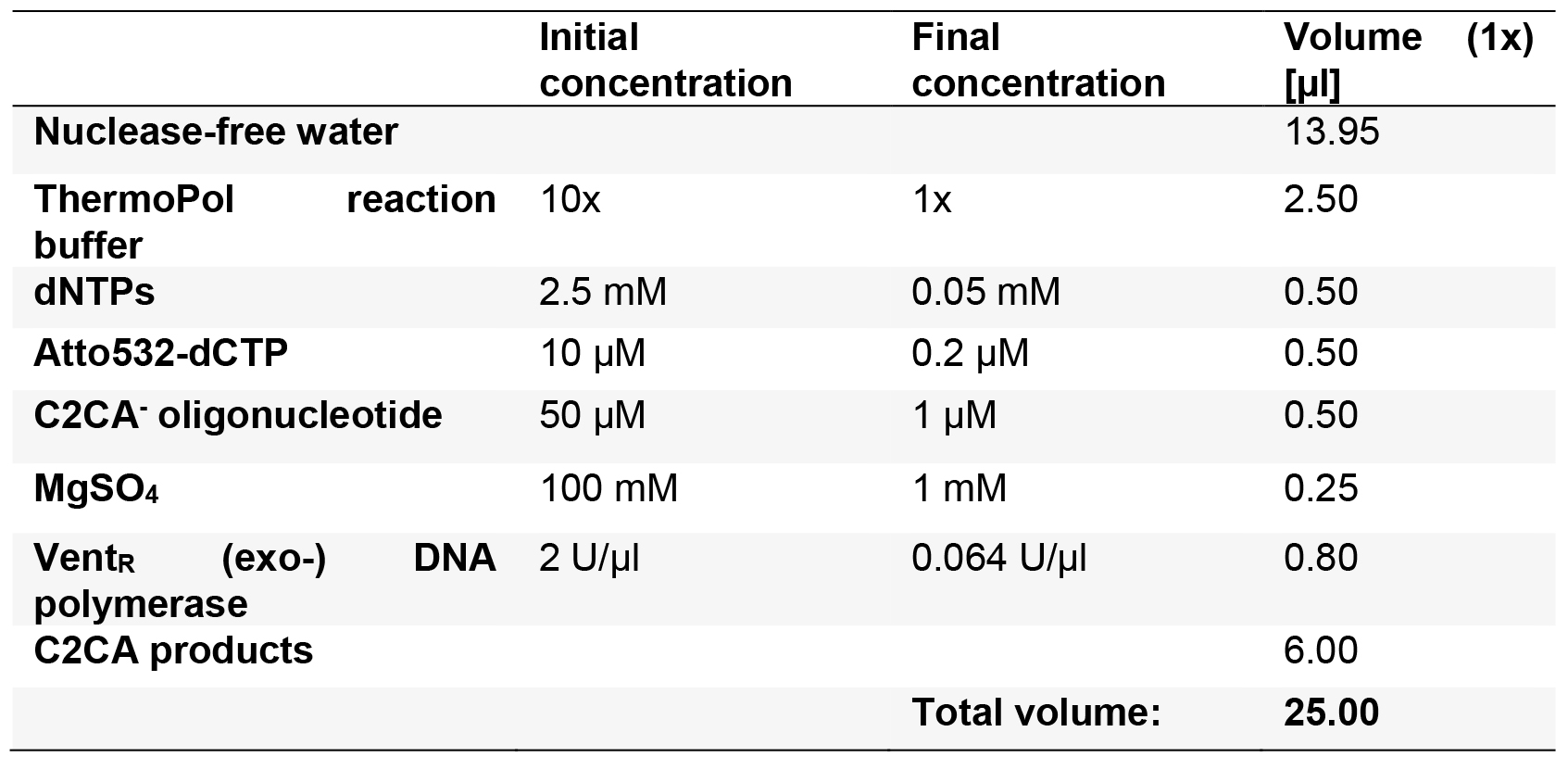
- Run the PCR program described in Table 4.
- Prepare the linear PCR master mix as described in Table 10 for n + 1 reactions and finally add 6 µl C2CA products.
- 33-plex pre-amplification PCR for β-lactamase-encoding gene identification
- Microarray analysis
- Prepare the detection mix for each sample by pooling 15 µl labelled PCR products, 15 µl labelled C2CA products, 1 µl Cy5-labelled ‘hybridization control’ oligonucleotide at a concentration of 50 µM and 30 µl ExpressHyb hybridization solution.
- Cover the microarray slide with a lifter slip (slides are not washed before use).
- Pipette the whole detection mix under the lifter slip.
- Put the slide in a humid incubation chamber (Note 12).
- Incubate at 65 °C for 45 min.
- Remove lifter slips.
- Wash slides in 2x SSC + 0.1% SDS for 5 min in a slide rack or staining jar while agitating.
- Wash slides in 0.2x SSC for 2 min in a slide rack or staining jar while agitating.
- Wash slides in H2O for 1 min in a slide rack or staining jar while agitating.
- Dry slides by centrifugation (2 min, 175 x g [900 rpm]) (Note 13).
- Scan the slides in a microarray scanner using the correct parameters for detection of Cy5 and Atto532 fluorophores.
- Prepare the detection mix for each sample by pooling 15 µl labelled PCR products, 15 µl labelled C2CA products, 1 µl Cy5-labelled ‘hybridization control’ oligonucleotide at a concentration of 50 µM and 30 µl ExpressHyb hybridization solution.
Data analysis
- The images for both fluorescence channels are imported into GenePix Pro 6.0.
- Create a .gal file to overlay an array containing the positions and names of your spotted microarray probes.
- Using the ‘Analyze’ function, mean and median intensities as well as standard deviations are calculated for the respective spots (and many more values).
- Export the results as text file and import them into Microsoft Excel or any other software suitable for data analysis (e.g., GraphPad Prism, R, IBM SPSS or other spreadsheet software).
Notes
- We use a Sartorius arium pro UV ultrapure water system. The lack of a hollow-fiber ultrafilter leads to incomplete removal of endotoxins, microorganisms, DNases and RNases, resulting in water which is not PCR-grade.
- Please consider that most enzymatic kits are not DNA-free. Enzymes such as polymerases are expressed and purified from cell cultures. In general, antibiotic resistance genes are used as selection markers and contamination of these and other genes may remain in the purified enzyme. Specialized kits have optimized purification procedures to remove such DNA contaminations.
- Microarray probe and primer design: The 5’-amino-modified microarray probes were designed with the ‘Probe Design’ function of the ARB software package. In summary, rRNA sequences were downloaded from GenBank and imported into ARB. Generated probes were optimized regarding maximum non-group hits, melting temperature, G+C content and minimum hairpin loops. Melting temperature and secondary structure were assessed using the ‘Oligo’ function of ARB. Finally, probes were manually optimized by adding or removing nucleotides regarding duplex formation and melting temperature. Using the ‘Probe Match’ function of ARB, the oligonucleotides were checked for their specificity. The multiplex PCR primers targeting the 33 beta-lactamase genes were designed using Primer3. When designing primers for multiplex PCR, it is important to avoid primer dimer formation and to have low differences in the primer melting temperature (here: 55 °C).
- Although not a requisite, we highly recommend to preparing oligonucleotide dilutions and Master mixes for PCR in a DNA/RNA-free environment to avoid cross-contamination. Genomic material (including PCR products) is added outside of the PCR hood to the enzymatic reactions. Sterilize the UV hood between different steps by UV irradiation.
- The preparation of genomic DNA extracts using bead-based cell disruption protocols has the advantage that genomic material of both Gram-positive and Gram-negative bacteria can be extracted simultaneously, while most commercially available kits require different protocols for these two types.
- Addition of antibiotics to the medium might be necessary because bacteria tend to lose their antibiotic resistance genes and plasmids due to reduced fitness and competition in the absence of antibiotics in liquid cell cultures.
- Bacterial or fungal growth is indicated by a cloudy haze. Bacteria should optimally be harvested at the end of the exponential phase (OD600 around 2).
- The phosphorylation of the 5’-end of the padlock probes is required in order to connect the 3’- with the 5’-end in the ligation reaction. We recommend ordering the padlock probes directly phosphorylated at the 5’-end. However, the cost of the oligonucleotides increases significantly if they are ordered with modifications.
- Spotting at low humidity causes the spots to dry too fast, resulting in torus-shaped spots. If the humidity is too high, spots become bigger and they might bleed into each other and create uneven, heterogeneous spots.
- The silylated aldehyde slides are delivered clean and there is no need for cleaning the slides before spotting. However, after clamping the glass slides into the mount, we recommend to blow off small splinters of glass with a high-pressure air pistol.
- In our case, 1 µl of genomic DNA extract corresponded to a concentration ranging between 11-48 ng µl-1.
- Be careful when handling the incubation chamber having water at the bottom. To avoid spilling water, put towels at the bottom, which still release humidity.
- Using our centrifuge (Heraeus 40R) and swinging bucket rotor, 900 rpm equal to 175 x g. Do not spin your slides at higher velocities to prevent damage to your slides or centrifuge.
Recipes
- CASO medium
Weight 30 g tryptic soy broth and dissolve in 1 L water
Autoclave for 15 min at 121 °C. Store at 4 °C - 1x phosphate-buffered saline (PBS)
Dilute 100 ml 10x PBS in 850 ml water
Adjust the pH using HCl or NaOH to 7.2, and adjust the final volume to 1 L
Store at room temperature - 2x spotting buffer (6x SSC, 3 M betaine)
Dissolve 20.27 g betaine monohydrate in 15 ml 20x SSC buffer
Adjust the volume to 50 ml with water
Sterile-filter the solution with a 0.20 µm syringe filter and transfer to a new 50 ml tube
Store at room temperature
Acknowledgments
This protocol has been adapted from Barišić et al. (2016), Journal of medical microbiology, 65(1), pp. 48-55.
References
- Barišić, I., Petzka, J., Schoenthaler, S., Vierlinger, K., Noehammer, C. and Wiesinger-Mayr, H. (2016). Multiplex characterization of human pathogens including species and antibiotic-resistance gene identification. J Med Microbiol 65(1): 48-55.
- Barišić, I., Schoenthaler, S., Ke, R., Nilsson, M., Noehammer, C. and Wiesinger-Mayr, H. (2013). Multiplex detection of antibiotic resistance genes using padlock probes. Diagn Microbiol Infect Dis 77(2): 118-125.
- Brandt, C., Braun, S.D., Stein, C., Slickers, P., Ehricht, R., Pletz, M.W. and Makarewicz, O., (2017). In silico serine β-lactamases analysis reveals a huge potential resistome in environmental and pathogenic species. Sci Rep 7.
- Bush, K., (2010). Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr Opin Microbiol 13(5): 558-564.
- Hall, B. G. and Barlow, M. (2003). Structure-based phylogenies of the serine β-lactamases. J Mol Evol 57(3): 255-260.
- Hall, B. G. and Barlow, M. (2004). Evolution of the serine β-lactamases: past, present and future. Drug Resist Updat 7(2): 111-123.
- Hardenbol, P., Yu, F., Belmont, J., Mackenzie, J., Bruckner, C., Brundage, T., Boudreau, A., Chow, S., Eberle, J., Erbilgin, A., Falkowski, M., Fitzgerald, R., Ghose, S., Iartchouk, O., Jain, M., Karlin-Neumann, G., Lu, X., Miao, X., Moore, B., Moorhead, M., Namsaraev, E., Pasternak, S., Prakash, E., Tran, K., Wang, Z., Jones, H. B., Davis, R. W., Willis, T. D. and Gibbs, R. A. (2005). Highly multiplexed molecular inversion probe genotyping: over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res 15(2): 269-275.
- Kong, K. F., Schneper, L. and Mathee, K. (2010). Beta‐lactam antibiotics: from antibiosis to resistance and bacteriology. APMIS 118(1) 1-36.
- Marik, P. E. (2014). Don’t miss the diagnosis of sepsis! Crit Care 18(5): 529.
- Mussap, M., Molinari, M. P., Senno, E., Gritti, P., Soro, B., Mannelli, S. and Fabris, C. (2007). New diagnostic tools for neonatal sepsis: the role of a real-time polymerase chain reaction for the early detection and identification of bacterial and fungal species in blood samples. J Chemother 19 Suppl 2: 31-34.
- Nilsson, M., Malmgren, H., Samiotaki, M., Kwiatkowski, M., Chowdhary, B. P. and Landegren, U. (1994). Padlock probes: circularizing oligonucleotides for localized DNA detection. Science 265(5181): 2085-2088.
- Wellinghausen, N., Kochem, A. J., Disque, C., Muhl, H., Gebert, S., Winter, J., Matten, J. and Sakka, S. G. (2009). Diagnosis of bacteremia in whole-blood samples by use of a commercial universal 16S rRNA gene-based PCR and sequence analysis. J Clin Microbiol 47(9): 2759-2765.
- Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., Aarestrup, F. M. and Larsen, M. V. (2012). Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67(11): 2640-2644.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Conzemius, R. and Barišić, I. (2017). Detection of Pathogens and Ampicillin-resistance Genes Using Multiplex Padlock Probes. Bio-protocol 7(16): e2504. DOI: 10.21769/BioProtoc.2504.
Category
Microbiology > Microbial genetics > DNA > PCR
Microbiology > Microbe-host interactions > Bacterium
Molecular Biology > DNA > Genotyping
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










