- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation and Analysis of Stromal Cell Populations from Mouse Lymph Nodes
Published: Vol 7, Iss 16, Aug 20, 2017 DOI: 10.21769/BioProtoc.2445 Views: 10980
Reviewed by: Ivan ZanoniMeenal SinhaAlesssandro Arduini

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Thrombopoietin-independent Megakaryocyte Differentiation of Hematopoietic Progenitor Cells from Patients with Myeloproliferative Neoplasms
Chloe A. L. Thompson-Peach [...] Daniel Thomas
Jan 20, 2023 2363 Views

Isolation, Purification, and Culture of Embryonic Melanoblasts from Green Fluorescent Protein–expressing Reporter Mice
Melissa Crawford [...] Lina Dagnino
Sep 5, 2023 1992 Views

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2243 Views
Abstract
Our protocol describes a simple procedure for isolating stromal cells from lymph nodes (LN). LN are disrupted then enzymatically digested with collagenase and dispase to produce a single cell suspension that can be stained with fluorescently labelled antibodies and analysed by flow cytometry. This protocol will enable identification of fibroblastic reticular cells (FRC), lymphatic endothelial cells (LEC), blood endothelial cells (BEC) as PNAd+ BEC that form LN high endothelial venules (HEV). This method can be applied to examine LN stromal cell responses during inflammatory events induced by infections or immunologic adjuvants and to subset most leukocytes found in LN.
Keywords: Lymphoid stromal cellsBackground
Lymph nodes (LN) are constructed of complex networks of mesenchymal and endothelial stromal cells. These include the fibroblastic reticular cells (FRCs), lymphatic endothelial cells (LECs) and blood endothelial cells (BECs). These stromal cells organize the complex microarchitecture of LN, enabling the support of immune cell migration, homeostasis, tolerance and cellular interactions required for the initiation of immune responses to pathogens and tumors. We have shown that the LN stromal cells can proliferate and expand in response to inflammatory signals and the recruitment of immune cells into LN that accompanies infections (Gregory et al., 2017). These stromal cells can also significantly modulate their transcriptional program to respond to infection, thereby supporting ongoing immune responses. This protocol enables reliable isolation of stromal cell subsets from LN both in the steady state and during disease. This enables phenotypic, functional, genetic or epigenetic investigation of LN stromal cells to reveal how they contribute to tissue homeostasis and immune responses.
Materials and Reagents
- 1,000 μl pipette tips (Neptune, catalog number: BT1250 )
- 200 μl pipette tips (Neptune, catalog number: BT200 )
- 20 μl pipette tips (Neptune, catalog number: BT20 )
- 10 μl pipette tips (Neptune, catalog number: BT10XLS3 )
- 24-well plate (Corning, catalog number: 3527 )
- 10 ml tubes (SARSTEDT, catalog number: 62.9924.284 )
- 50 ml tubes (Greiner Bio One International, catalog number: 227261 )
- Nylon mesh 70 μm (Clear Edge, catalog number: PA75-1750 )
- 5 ml polystyrene round bottom tube (Corning, Falcon®, catalog number: 352008 )
- 96-well round-bottom plate (Corning, catalog number: 3788 )
- Spatula (VWR, catalog number: 10806-408 )
- C57BL\6 mice (THE JACKSON LABORATORY, catalog number: 000664 )
- Blank calibration particles (BD, BD Biosciences, catalog number: 556296 )
- HEPES (Thermo Fisher Scientific, GibcoTM, catalog number: 11344033 )
- L-Glutamine (Thermo Fisher Scientific, GibcoTM, catalog number: 21051024 )
- Benzylpenicillin (CSL, catalog number: AUST R 10329 )
- Streptomycin sulphate (Thermo Fisher Scientific, GibcoTM, catalog number: 11860038 )
- 2-Mercaptoethanol (Sigma-Aldrich, catalog number: M6250 )
- RPMI (Thermo Fisher Scientific, GibcoTM, catalog number: 11875093 )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10099141 )
- Collagenase D (Sigma-Aldrich, catalog number: 11088882001 )
Manufacturer: Roche Diagnostics, catalog number: 11088882001 . - Dispase II (Sigma-Aldrich, catalog number: 04942078001 )
Manufacturer: Roche Diagnostics, catalog number: 04942078001 . - DNase I (Sigma-Aldrich, catalog number: DN25-1G )
- Phosphate-buffered saline (PBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10010023 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A7906-500G )
- Ethylenediaminetetraacetic acid disodium salt dihydrate (EDTA) (Sigma-Aldrich, catalog number: E5134-1KG )
- Antibodies
- gp38-PE, 8.1.1 (BioLgend, catalog number: 127408 )
- CD31-PE-Cy7, 390 (BioLegend, catalog number: 102524 )
- CD45.2-A700, 104 (BioLegend, catalog number: 109822 )
- Ter-119-A700, TER-119 (BioLegend, catalog number: 116220 )
- HEV-A488, MECA-79 (Thermo Fisher Scientific, eBioscienceTM, catalog number: 53-6036-82 )
- CD16/32-purified, 2.4G2 (BD, BD Biosciences, catalog number: 553142 )
- gp38-PE, 8.1.1 (BioLgend, catalog number: 127408 )
- Live/Dead Fixable Near-IR cell stain kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: L10119 )
- Supplementum Completum (SC) (see Recipes)
- RP-2 solution (see Recipes)
- Digestion mix 1 (see Recipes)
- Digestion mix 2 (see Recipes)
- FACS buffer (see Recipes)
- Lymph node stromal cell antibody cocktail (see Recipes)
Equipment
- Water bath (VWR, catalog number: 89501-464 )
- Dumont No.5 forceps (Fine Science Tools, catalog number: 11251-10 )
- Graefe forceps (Roboz Surgical Instrument, catalog number: RS-5137 )
- Analytical balance for weighing (Ohaus, catalog number: 30208454 )
- 1,000 μl pipette (Eppendorf, catalog number: 3121000120 )
- 200 μl pipette (Eppendorf, catalog number: 3121000082 )
- 20 μl pipette (Eppendorf, catalog number: 3121000040 )
- 10 μl pipette (Eppendorf, catalog number: 3121000015 )
- Refrigerated centrifuge with plate carriers (Beckman Coulter, model: Allegra® X-15R , catalog number: B09822; Beckman Coulter, catalog number: 368914 )
- Flow cytometer (BD, BD Biosciences, model: BD LSRFORTESSA )
Software
- FlowJo (FlowJo)
- Prism (GraphPad)
Procedure
- Isolation and digestion of lymph nodes
- Prepare digestion mixes 1 and 2 (see Recipes) and place in a water bath at 37 °C.
Note: The volume of the digestion mixes prepared is dependent on the number of samples required for the experiment. 1 ml of each digestion mix is required for up to 6 skin draining lymph nodes (LN). Digestion mixes are prepared fresh for each experiment. We prepare the required amount of each enzyme for each experiment. However, users can prepare a 100x stock of the enzymes (in RPMI) and store them at -20 °C for further usage if preferred. - Harvest LN from mice, place in wells of a 24-well plate containing 1 ml of RP-2 solution per well (see Recipes) and keep on ice.
Note: We typically process two skin draining LN from non-infected mice (inguinal, axillary or brachial). During an inflammatory episode such as Herpes Simplex Virus (HSV) flank infection, we process the brachial LN individually. If popliteal LN are isolated, we pool and process at least 4 LN from non-infected mice, and only one during an inflammatory event.
To assist with the identification of mouse LN, users can refer to a diagram of the localization of LN in the mouse model, available here:
http://www.informatics.jax.org/greenbook/figures/figure13-4.shtml - Take the LN out of the plate and disrupt the LN into 4-5 pieces with two thin forceps.
- Transfer disrupted LN into a 10 ml tube and add 1 ml of digest mix 1.
Note: During this step, LN are not put back into the 24-well plate. Thus they are digested in 1 ml with the digestion mix 1. - Place tubes in the water bath and incubate for 25 min at 37 °C.
- Invert tube and flick a few times during digestion.
- After incubation, cut off the tip of 1 ml pipette and thoroughly mix the digested LN for 30 sec, up to 1 min. Leave undisturbed until remaining visible pieces of tissue settle to the bottom of the tube.
- Remove supernatant without taking the undigested pieces of LN and transfer into a 50 ml tube containing 15 ml of FACS buffer (see Recipes) and keep on ice.
Note: Make sure to remove as much supernatant as possible without taking any pieces of LN. There should be minimal medium left in the tube. One round of digestion is not enough to properly digest LN and thus two rounds of digestion are necessary to maximize the yield of SC to be released. - Add 1 ml of digestion mix 2 to the undigested pieces of LN left from step A7 and place in the water bath for 15 min at 37 °C.
- After incubation, thoroughly mix the digested LN with 1 ml pipette for 1 min.
- Transfer supernatant to tubes containing the cell suspension from step A8.
Note: LN should completely be digested after this step. If any pieces are remaining, repeat steps A9 and A10 with 10-min incubation only. - Centrifuge cell suspension for 5 min at 500 x g, 4 °C.
- Aspirate supernatant and resuspend cell suspension in 500 (for up to 6 skin draining LN, or popliteal LN) or 1,000 μl of FACS buffer (if more than 6 skin draining LN).
- Filter cell suspension through 70 μm nylon mesh in a 5 ml FACS tube and count cells.
Note: Users can also use counting beads (blank calibration particles) during acquisition by flow cytometry, as per the manufacturer’s instructions.
- Prepare digestion mixes 1 and 2 (see Recipes) and place in a water bath at 37 °C.
- Antibody staining of LN cell suspensions
- Transfer up to 200 μl of the cell suspension into each well of a 96 U-bottom well plate.
Note: If plate holders are not available, the staining can be performed in small Eppendorf tubes and a refrigerated micro-centrifuged can be used. Alternatively, staining can be performed in FACS tubes. - Centrifuge plate at 500 x g for 5 min and at 4 °C then flick off the supernatant.
- Add 50 μl of the stromal cell antibody cocktail mix (see Recipes) to each well, resuspend the cells and incubate for 25 min at 4 °C in the dark.
- Add 150 μl of FACS buffer per well, centrifuge the plate and flick off the supernatant.
- Resuspend cell suspension in 200 μl of FACS buffer and run the stained cells in a flow cytometer (BD LSRFORTESSA).
- Transfer up to 200 μl of the cell suspension into each well of a 96 U-bottom well plate.
Data analysis
It is recommended to include 4-5 mice per group and perform 2-3 independent experiments. Flow cytometry files are analysed using FlowJo to gate and quantitate individual stromal cells subsets (Figure 1). To analyse LN stromal cell subsets during infection, draining and non-draining LN are compared, and populations of stromal cells quantitated and numbers plotted (Figure 2). This procedure will also release all leukocytes from the LN, enabling additional antibody staining and analysis of other cell types if required (see Note 1). To assess statistical significance between groups, unpaired t-tests or Mann-Whitney tests should be performed, as appropriate, using Prism (GraphPad).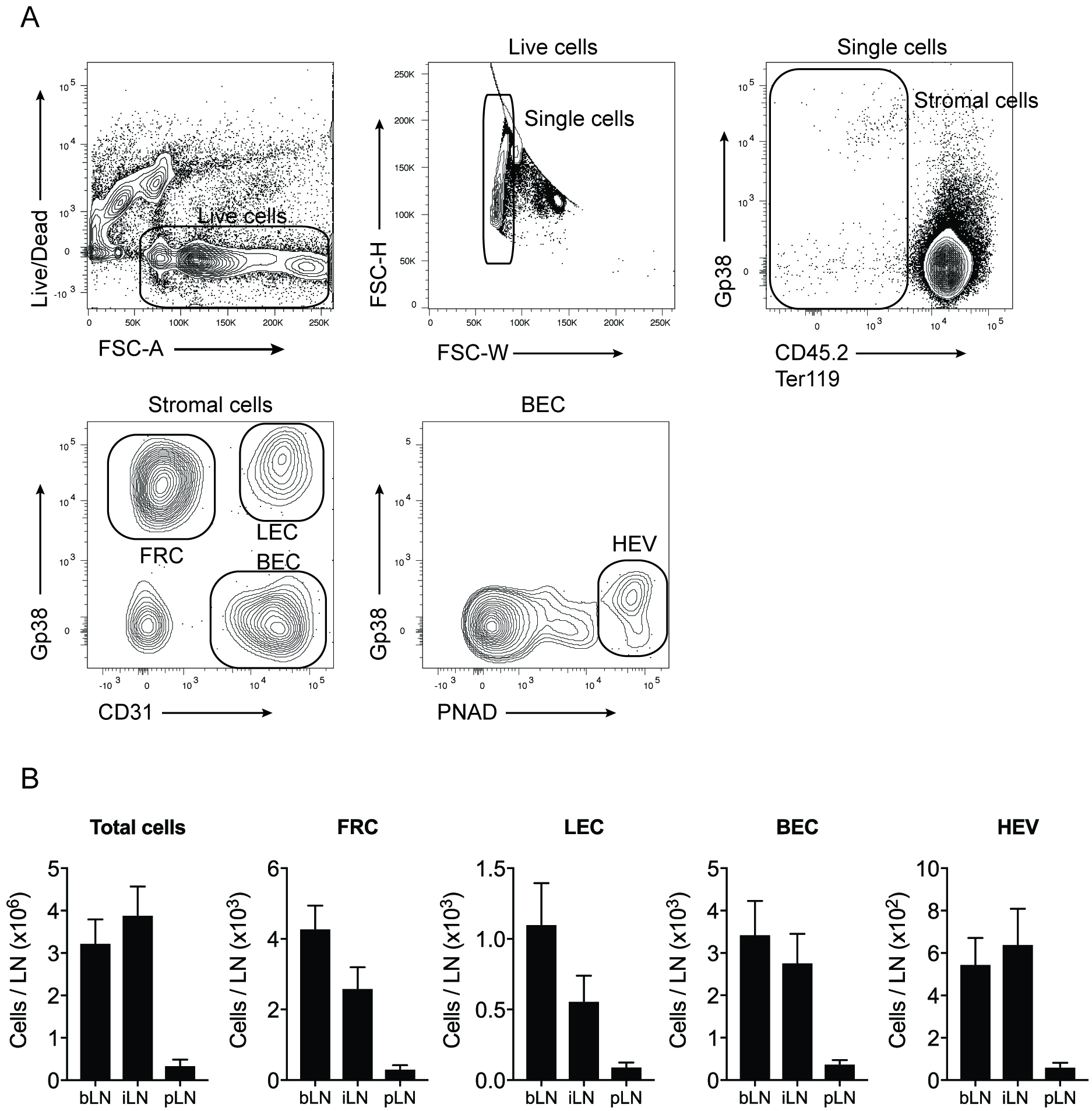
Figure 1. Gating strategy used to identify LN stromal cell subsets. Dead cells and doublet cells are first excluded. Stromal cells lack CD45.2 and Ter119, markers of hematopoietic cells and erythroid cells, respectively. The use of gp38 and CD31 allows the identification of fibroblastic reticular cells (FRC), lymphatic endothelial cells (LEC) and blood endothelial cells (BEC). The marker PNAD is used to identify the BEC that form the high endothelial venules (HEV) in LN. Bar graphs show the quantification of the stromal cell subsets in the brachial LN (bLN), inguinal LN (iLN) and the popliteal LN (pLN) at steady state.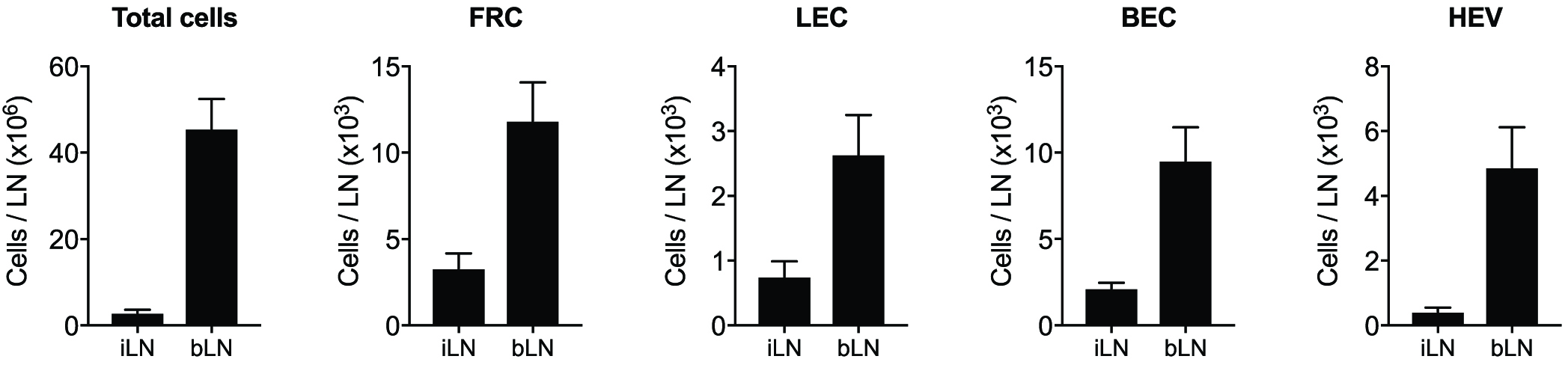
Figure 2. Analysis of stromal cell subsets in draining brachial LN and non-draining inguinal LN by flow cytometry after Herpes simplex virus (HSV) infection. Mice were infected by scarification on the left flank skin with HSV and LN harvested 7 days post-infection for stromal cell analysis. During an immune challenge, stromal cells expand to accommodate the increase of total cellularity in the LN.
Notes
Dispase treatment can cleave certain cell surface markers. We found that typical markers of stromal cells are not affected by our enzymatic digestion method. We routinely analyse leukocyte subsets in the LN and found that CD8α and CD19 staining is diminished when this protocol is applied. The use of CD8β and B220 as alternative markers is recommended.
Recipes
- Supplementum Completum (SC)
For 1 L solution:
23.83 g HEPES
6 g L-glutamine
2 vials benzylpenicillin
2 g streptomycin sulphate
70 μl 2-mercaptoethanol
1 L RPMI
Filter, aliquot in 25 ml solution into 50 ml tubes and keep at -20 °C - RP-2 solution
For 500 ml solution:
500 ml of RPMI
10 ml decomplemented fetal bovine serum
20 ml of SC solution
Note: Keep at 4 °C, no longer than 2 months. - Digestion mix 1 (Collagenase D: 1 mg/ml, DNase I: 0.1 mg/ml)
For 20 ml solution:
20 ml RP-2 solution
20 mg collagenase D
2 mg DNase I
Note: Discard at the end of the experiment. - Digestion mix 2 (Collagenase D: 1 mg/ml, DNase I: 0.1 mg/ml, Dispase II 0.8 mg/ml)
For 20 ml solution:
20 ml RP-2 solution
20 mg collagenase D
2 mg DNase I
16 mg dispase II
Note: Discard at the end of the experiment. - FACS buffer
For 500 ml solution:
500 ml of PBS
2.5 g of BSA
5 ml 0.5 M EDTA
Note: Keep at 4 °C, no longer than 3 months. - Lymph node stromal cell antibody cocktail
For 500 μl solution:
500 μl FACS buffer
1 μl gp38-PE (1/500)
1 μl CD31-PE-Cy7 (1/500)
10 μl CD45.2-A700 (1/50)
10 μl Ter119-A700 (1/50)
2.5 μl PNAd-A488 (1/200)
1 μl Live/Dead cell marker-NearIR (1/500)
2.5 μl purified CD16/CD32 (1/200)
Note: Keep at 4 °C or on ice.
Acknowledgments
This protocol was adapted from our publication (Gregory et al., 2017). This work was supported by the Australian Research Council.
Competing interests
The authors declare that they have no competing interests.
References
- Gregory, J. L., Walter, A., Alexandre, Y. O., Hor, J. L., Liu, R., Ma, J. Z., Devi, S., Tokuda, N., Owada, Y., Mackay, L. K., Smyth, G. K., Heath, W. R. and Mueller, S. N. (2017). Infection programs sustained lymphoid stromal cell responses and shapes lymph node remodeling upon secondary challenge. Cell Rep 18(2): 406-418.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Alexandre, Y. O. and Mueller, S. N. (2017). Isolation and Analysis of Stromal Cell Populations from Mouse Lymph Nodes. Bio-protocol 7(16): e2445. DOI: 10.21769/BioProtoc.2445.
Category
Cell Biology > Cell isolation and culture > Cell isolation > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










