- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation of Cytosol, Microsome, Free Polysomes (FPs) and Membrane-bound Polysomes (MBPs) from Arabidopsis Seedlings
Published: Vol 7, Iss 15, Aug 5, 2017 DOI: 10.21769/BioProtoc.2436 Views: 12717
Reviewed by: Arsalan DaudiLiping ShenWenrong He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Optimized Isolation of Lysosome-Related Organelles from Stationary Phase and Iron-Overloaded Chlamydomonas reinhardtii Cells
Jiling Li and Huan Long
Nov 20, 2024 1766 Views

Isolation and Biophysical Characterization of Extracellular Vesicles From Hairy Root Cultures
Marisa Conte [...] Alfredo Ambrosone
Mar 5, 2025 2218 Views

Rapid Miniprep of Intact Chloroplasts from Arabidopsis thaliana Leaves
Brenda A. Carranza-Correa [...] Manuel Gutiérrez-Aguilar
May 20, 2025 2645 Views
Abstract
The plant endomembrane system plays vital roles for synthesis, modification and secretion of proteins and lipids. From the classic view, only mRNAs encoding secreted proteins could be targeted to the endoplasmic reticulum (ER) for translation via a co-translational translocation manner, however, recently this model has been challenged by accumulative evidence that lots of cytosolic mRNAs could also associate with ER, and that some categories of small RNAs are enriched on ER. These results suggested unrevealed functions of ER beyond our current knowledge. The large scale identification of RNAs and proteins on microsome is crucial to demonstrating the ER function and the studies will be boosted by next generation sequencing technology. This protocol provides a technical workflow to isolate the cytosol, microsome, free polysome (FP) and membrane bound polysome (MBP) from plant tissue. The isolated fractions are suitable for genome wide profiling of mRNAs, small RNAs and proteins.
Keywords: CytosolBackground
Plant endomembrane system is very important for cell wall formation, lipid biosynthesis, protein synthesis, modification, folding and trafficking. According to the co-translational translocation model, signal peptides at the N-terminal of secreted proteins are synthesized by cytosolic polysomes, and then recognized by signal recognition particles on ER, and the remaining portion of proteins will be subsequently synthesized on ER. According to this model, only mRNAs encoding for secreted proteins could be brought to ER for translation (Peter and Johnson, 1994). However, large portion of mRNAs were identified from mammalian and plant cell ERs (Lerner et al., 2003; de Jong et al., 2006), and recent studies revealed that ER also functions as a key hub for small RNA function in plant (Li et al., 2013 and 2016). These findings broadened our knowledge about ER functionality. Large scale identification of mRNAs, small RNAs and proteins from ER of cells upon different developmental stages and environmental stimuli will provide valuable clues for elucidating new functions of ER. Here, we describe a protocol to isolate the cytosol, microsome, FP and MBP from Arabidopsis thaliana, and it could be adapted to rice, maize and other plants.
Materials and Reagents
- Pipette tip (Denville Scientific, catalog numbers: P2101 , P2102 , P2109 ), autoclave before use
- 50 ml tube
- Miracloth (EMD Millipore, catalog number: 475855-1R )
- 15 ml tube
- 13 x 51 mm centrifuge tubes (Beckman Coulter, catalog number: 326819 )
- 25 x 89 mm centrifuge tubes (Beckman Coulter, catalog number: 355631 )
- Arabidopsis ecotype Columbia-0 maintained by our own laboratory
- Murashige and Skoog medium
- Liquid nitrogen
- 1% (v/v) Triton X-100
- DEPC H2O
- Tris base (Fisher Scientific, catalog number: BP152-5 )
- Hydrochloric acid (HCl) (Fisher Scientific, catalog number: A142-212 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9333 )
- MgOAc
- Ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid (EGTA) (Sigma-Aldrich, catalog number: E3889 )
- Sucrose (Fisher Scientific, catalog number: BP220-212 )
- Dithiothreitol (DTT) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0861 )
- Cycloheximide (Sigma-Aldrich, catalog number: C1988 )
- Chloramphenicol (Sigma-Aldrich, catalog number: C0378 )
- Ethanol
- SUPERaseIN (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM2696 )
- Magnesium acetate tetrahydrate (MgCl2·4H2O) (Sigma-Aldrich, catalog number: M5661 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Fisher Scientific, catalog number: BP214-500 )
- Proteinase inhibitor cocktail-EDTA free (Roche Diagnostics, catalog number: 18970600 )
- Ribosome extraction buffer (see Recipes)
- Sucrose cushion buffer (see Recipes)
- Resuspending buffers (see Recipes)
- 10x sucrose salt (for 15-60% sucrose gradient column) (see Recipes and Notes)
Equipment
- Pipette (Eppendorf)
- Plant growth chamber (Percival Scientific, model: CU-36L4 )
- L8-70M Ultracentrifuge (Beckman Coulter, model: L8-70M )
- SW 28 rotor (Beckman Coulter, model: SW 28 Ti )
- SW 55 Ti rotor (Beckman Coulter, model: SW 55 Ti )
- Type70 Ti rotor (Beckman Coulter, model: Type70 Ti )
- 25 x 89 mm bottle, with cap assembly (Beckman Coulter, catalog number: 355618 )
- Vacuum pump
- Centrifuge (Eppendorf, model: 5424 R )
- High speed centrifuge (Beckman Coulter, model: Avanti J-E Series )
- NanoDrop spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDropTM 2000 )
- Density gradient fractionation systems (BRANDEL, model: BR-188 )
- 37 °C incubator
Software
- Data acquisition software (Brandel, model: PEAK CHART)
Procedure
This protocol allows the simultaneous isolation of FP and MBP from the same plant sample. Briefly, the cytosol and microsome fractions are separated by centrifugation, and microsome fraction is dissolved with extraction buffer supplemented with detergent. Both cytosol and microsome lysates are passed through sucrose cushion solution by ultracentrifugation to obtain FP and MBP pellets which are subsequently subjected to density gradient fractionation and profile analyses (Figure 1).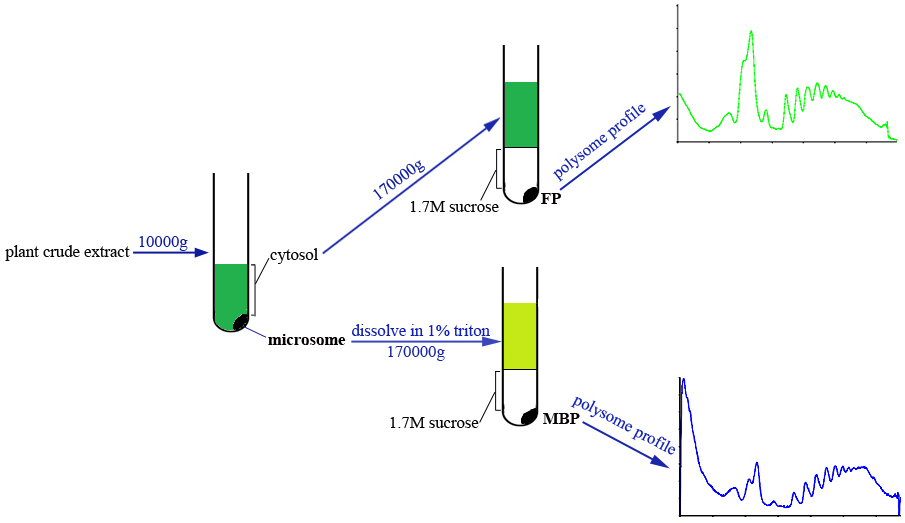
Figure 1. Scheme of FP and MBP isolation
- Col-0 seeds are sterilized and plated on Murashige and Skoog medium, and plants are grown in a growth chamber at 23 °C under 16 h light/8 h dark cycles for 12 days.
- 2 g seedlings are ground into fine powder in liquid nitrogen, and are suspended in 8 ml ribosome extraction buffer (see Recipes) in a 50 ml tube. Keep on ice for 20 min.
- The slurry is filtered with 2 layers of Miracloth to a 15 ml tube and centrifuged twice at 10,000 x g for 10 min to remove the debris.
- The supernatant is transferred into a Beckman centrifuge tube and centrifuged at 30,000 x g for 30 min with a Beckman SW28 rotor. Transfer the supernatant to a new tube as the cytosol fraction and keep it on ice.
- Resuspend the pellet with 8 ml ribosome extraction buffer followed by centrifugation at 30,000 x g for another 30 min. Discard the supernatant, and the pellet is kept as the microsome fraction.
Note: The cytosol and microsome fractions from steps 3 and 4 are ready for RNA and protein extraction. If you want to perform FP/MBP isolation, please continue the following steps. - Dissolve the microsome pellet with 8 ml ribosome extraction buffer supplemented with 1% (v/v) Triton X-100. Keep it on ice for 20 min.
- Subject the cytosol extract (step 3) and microsome lysate (step 5) to centrifugation at 30,000 x g for 30 min with a Beckman SW28 rotor to remove any residual membranes.
- Transfer 8 ml sucrose cushion solution (see Recipes) into a centrifugation bottle (Beckman centrifuge) suitable for Type70 Ti rotor (Beckman Coulter), and then slowly load the clarified cytosol or microsome lysate from step 6 on the top of the sucrose cushion.
Note: Be careful not to disturb the sucrose cushion layer. - Centrifuge at 183,960 x g with Type70 Ti rotor (Beckman Coulter) at 4 °C for 3 h.
- Draw a circle around the ribosome pellet with a marker pen, and remove all liquid in the tube with a pipette or a vacuum pump (Video 1). Hold the tube with the marked position upward, and carefully wash the inner surface of the tube except for the marked area by 1 ml ddH2O three times with a pipette (Video 1). The purpose of this step is to remove the residual salt and sucrose in the tube. Any touching with the FP/MBP pellet either by pipette tip or water must be avoided.Video 1. Removal of supernatant by vacuum and washing of the tube inner wall
- Resuspend the pellets in 400 μl resuspension buffer (see Recipes), and transfer them to nuclease free microcentrifuge tubes. Keep the tubes on ice for 30 min.
- Centrifuge at 16,000 x g for 5 min at 4 °C to remove debris, and transfer the supernatants to new tubes.
Note: The samples obtained from step 11 are ready for RNA and protein extraction of FP/MBP. If you want to check the FP/MBP profiles, continue the following steps. - Measure the OD260 of the samples from step 11 with NanoDrop spectrophotometer.
- Slowly load 1,000 OD260 of FP or 200 OD260 of MBP on the top of 15-60% sucrose gradient column (see Notes). The yield of MBP is much lower than FP, but 200 OD260 is enough for the MBP profile analysis.
Note: Be careful not to disturb the sucrose gradient. It is important to keep the pipette tip and the surface of the gradient solution nicely touched (but not protruding into the solution) during the loading, otherwise droplets may be formed and the gradient will be disturbed. - Centrifuge at 237,020 x g with SW55 Ti rotor (Beckman Coulter) for 1.5 h at 4 °C.
- Perform the density gradient fractionation. The fractionation system is composed of a syringe pump, a tube piercer stand, a detector, a fraction collector and the Peakchart software (Figure 2A). The gradient column is mounted onto the tube piercer stand and is pierced by the needle at the bottom of stand (see Video 2). For the syringe pump, put the speed mode switch to ‘normal’ position, and the fluid direction switch to ‘off’ position; Turn the control mode knob to ‘remote start/stop’, and adjust the fluid speed to 1.5 ml/min (Figure 2B); For the UA-6 detector, set the sensitivity value as ‘1’, and the chart speed as ‘150 cm/h’ (Figure 2C). The system was under control of the Peakchart software, and ribosome profiles were recorded by the software and the detector (Figure 2D).
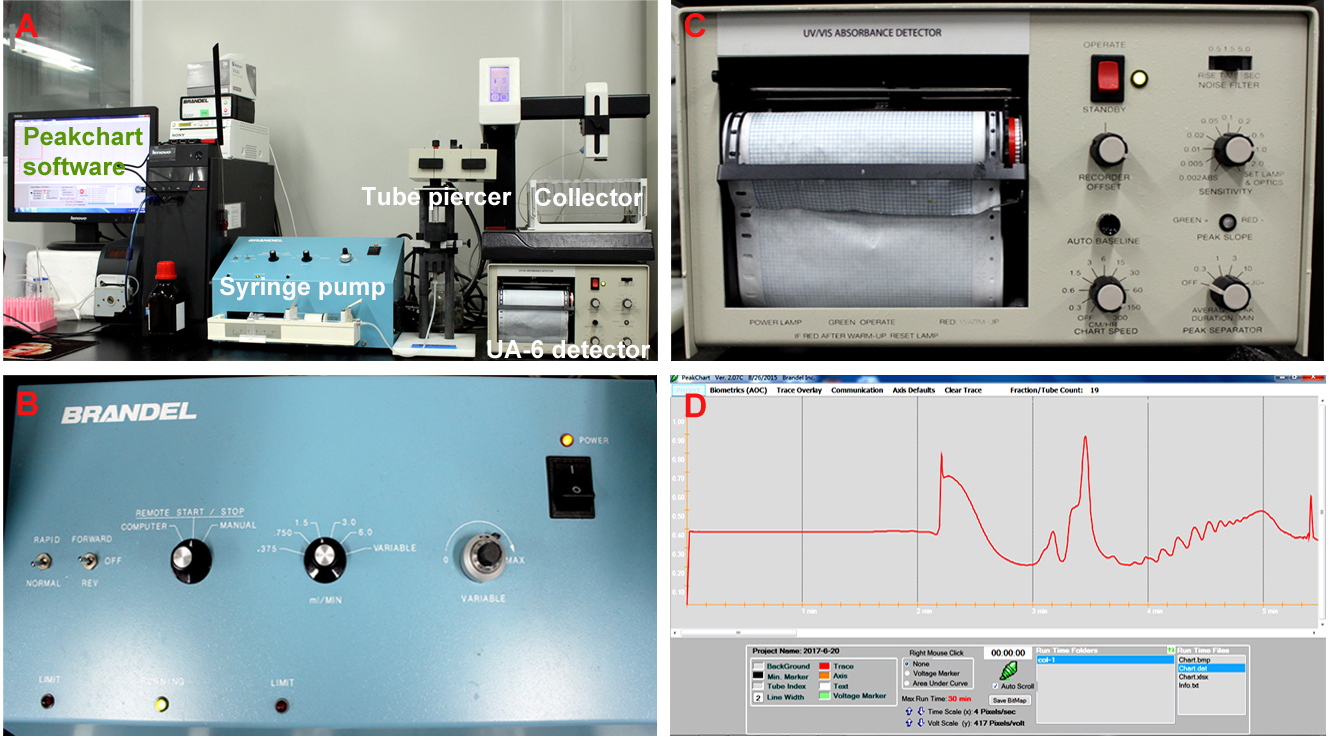
Figure 2. The density gradient fractionation system. A. The overview of the fractionation system. The gradient column is attached to the tube piercer stand, and pierced by the needle at the bottom of the stand. The gradient solution is slowly pushed out from the top of the column by the chase fluid in the syringe pump, and A254 nm absorbance was recorded by the UA-6 detector and the Peakchart software. B. The front panel of the syringe pump. The positions of the switches and knobs reflect the parameter settings during fractionation. C. The front panel of the UA-6 detector. The positions of the knobs reflect the parameter settings during fractionation. D. A screenshot of the Peakchart software. The start or stop of the entire system is controlled by the green button at the bottom center, the profile is shown on the screen in real time manner, and the data are automatically saved when the procedure completes.Video 2. Attachment of the gradient column to the density gradient fractionation system - Analyze the ribosome profile. The typical FP/MBP profiles are shown in Figure 3. The different peaks represent 40S small subunit, 60S large subunit, the 80S monoribosome and polyribosomes respectively, and a good isolation of FP or MBP should display a profile with these distinct peaks and a hill shaped pattern instead of a decline curve in the polysome region. Note that the peak of 80S monomer of MBP is much lower than that of FP, and 60S and 80S fractions were usually combined in MBP.
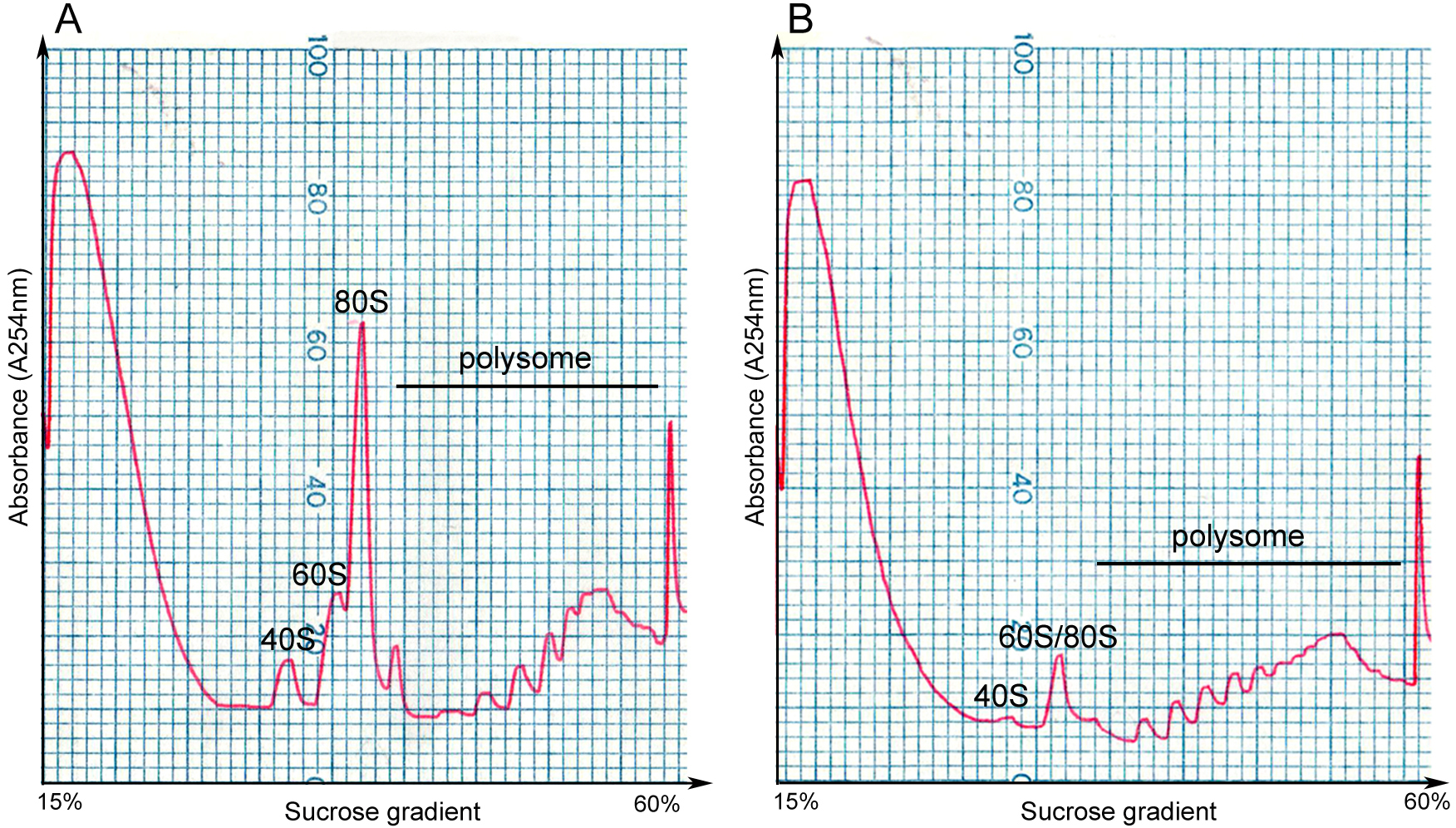
Figure 3. Profiles of FP and MBP. FP (A) and MBP (B) are separated in 15-60% sucrose gradient by ultracentrifugation, and are fractionated by gradient fractionation system subsequently. The x-axis indicates the sucrose concentration in the corresponding gradient, and the y-axis represents the absorbance level at 254 nm. 40S: small subunit of ribosome; 60S: large subunit of ribosome; 80S: the monoribosome complex.
Notes
Preparation of 15-60% sucrose gradient column. All stock solutions are prepared with DEPC H2O except for CHX and CHL which were prepared with ethanol.
- Prepare sucrose solutions with different sucrose concentration (for 10 gradient columns, Table 1):
Table 1. The recipes for preparing the 15-60% sucrose gradient column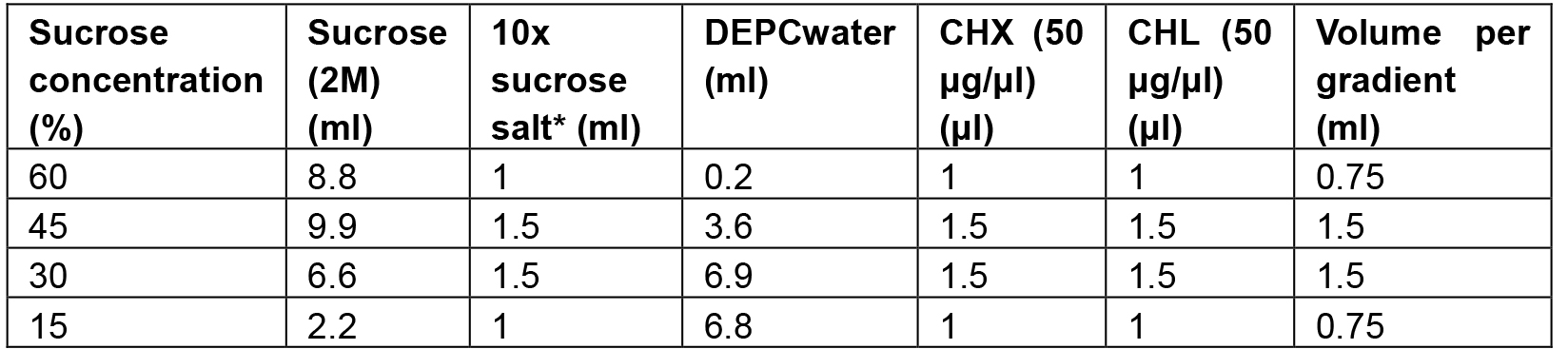
- Place 13 x 51 mm centrifuge tubes (Beckman) into a rack that can withstand -80 °C
- Start with the 60% sucrose layer, pipette 0.75 ml 60% sucrose solution into a 13 x 51 mm centrifuge tube (Beckman), avoiding any air bubbles, and then freeze for 1h at -80 °C.
- Add the next gradient layer with the volumes indicated in the table, freeze again, and continue with the last two layers.
- Store the sucrose gradient columns at -80 °C. The gradient columns could be used in 3 months if they are stored properly.
- Before use, remove the column from the freezer, and thaw in a 37 °C incubator for exactly 1 h followed by cooling down in a cold room or refrigerator for another 1 h.
Recipes
- Ribosome extraction buffer
0.2 M Tris-HCl, pH 8.5
0.1 M KCI
70 mM MgOAc
50 mM EGTA
0.25 M sucrose
10 mM DTT
50 μg/ml Cycloheximide (CHX) (stock 50 μg/μl in ethanol)
50 μg/ml Chloramphenicol (CHL) (stock 50 μg/μl in ethanol)
2.5 U/ml SUPERaseIN - Sucrose cushion solution
0.4 M Tris-HCl, pH 9.0
0.2 M KCI
0.005 M EGTA
0.035 M MgCl2
1.75 M sucrose
5 mM DTT
50 μg/ml CHX (stock 50 μg/μl in ethanol)
50 μg/ml CHL (stock 50 μg/μl in ethanol) - Resuspension buffer
0.2 M Tris-HCl, pH 9.0
0.2 M KCl
0.025 M EGTA
0.035 M MgCl2
5 mM DTT
50 μg/ml CHX (stock 50 μg/μl in ethanol)
50 μg/ml CHL (stock 50 μg/μl in ethanol) - 10x sucrose salt (for 15-60% sucrose gradient column)
0.4 M Tris-HCl, pH 8.4
0.2 M KCI
0.1 M MgCI2
Note: All buffers were prepared with DEPC water if not emphasized; DTT, CHX, CHL and SUPERaseIN need be added freshly.
Acknowledgments
This protocol was adapted from our previous work (Li et al., 2016). We thank Dr. Xuemei Chen for suggestions on this protocol. The work was supported by grants from the science technology and innovation committee of Shenzhen municipality (JCYJ20151116155209176, KQCX2015033110464302, KY20150114), and the Key Laboratory of Shenzhen (ZDSYS20141118170111640).
References
- de Jong, M., van Breukelen, B., Wittink, F. R., Menke, F. L., Weisbeek, P. J. and Van den Ackerveken, G. (2006). Membrane-associated transcripts in Arabidopsis; their isolation and characterization by DNA microarray analysis and bioinformatics. Plant J 46(4): 708-721.
- Lerner, R. S., Seiser, R. M., Zheng, T., Lager, P. J., Reedy, M. C., Keene, J. D. and Nicchitta, C. V. (2003). Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA 9(9): 1123-1137.
- Li, S., Le, B., Ma, X., Li, S., You, C., Yu, Y., Zhang, B., Liu, L., Gao, L., Shi, T., Zhao, Y., Mo, B., Cao, X. and Chen, X. (2016). Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. Elife 5.
- Li, S., Liu, L., Zhuang, X., Yu, Y., Liu, X., Cui, X., Ji, L., Pan, Z., Cao, X., Mo, B., Zhang, F., Raikhel, N., Jiang, L. and Chen, X. (2013). MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153(3): 562-574.
- Peter, W. and Johnson, A. E. (1994). Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol 10: 87-119.
Article Information
Copyright
Zhao and Li. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Zhao, Y. and Li, S. (2017). Isolation of Cytosol, Microsome, Free Polysomes (FPs) and Membrane-bound Polysomes (MBPs) from Arabidopsis Seedlings. Bio-protocol 7(15): e2436. DOI: 10.21769/BioProtoc.2436.
- Li, S., Le, B., Ma, X., Li, S., You, C., Yu, Y., Zhang, B., Liu, L., Gao, L., Shi, T., Zhao, Y., Mo, B., Cao, X. and Chen, X. (2016). Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. Elife 5.
Category
Plant Science > Plant cell biology > Organelle isolation
Cell Biology > Organelle isolation > Microsome
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












