- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analyzing the Properties of Murine Intestinal Mucins by Electrophoresis and Histology
Published: Vol 7, Iss 14, Jul 20, 2017 DOI: 10.21769/BioProtoc.2394 Views: 20583
Reviewed by: Andrea PuharDavid A. CisnerosAna Santos Almeida

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measuring Protein Synthesis during Cell Cycle by Azidohomoalanine (AHA) Labeling and Flow Cytometric Analysis
Koshi Imami and Tomoharu Yasuda
Apr 20, 2019 9752 Views

Analysis of the Ubiquitination and Phosphorylation of Vangl Proteins
Di Feng [...] Bo Gao
Oct 20, 2022 3422 Views

Isoform-specific, Semi-quantitative Determination of Highly Homologous Protein Levels via CRISPR-Cas9-mediated HiBiT Tagging
Kristina Seiler [...] Mario P. Tschan
Jul 20, 2023 2521 Views
Abstract
Specialized secretory cells known as goblet cells in the intestine and respiratory epithelium are responsible for the secretion of mucins. Mucins are large heavily glycosylated proteins and typically have a molecular mass higher than 106 Da. These large proteins are densely substituted with short glycan chains, which have many important functional roles including determining the hydration and viscoelastic properties of the mucus gel that lines and protects the intestinal epithelium. In this protocol, we comprehensively describe the method for extraction of murine mucus and its analysis by agarose gel electrophoresis. Additionally we describe the use of High Iron Diamine-Alcian Blue, Periodic Acid Schiff’s-Alcian Blue and immune–staining methods to identify and differentiate between the different states of glycosylation on these mucin glycoproteins, in particular with a focus on sulphation and sialylation.
Keywords: SialylationBackground
A layer of mucus protects the intestinal epithelium and primarily consists of mucins, water, proteins and inorganic salts. The viscous and gel-like properties of the mucus barrier, which enable it to physically protect and lubricate the mucous membranes, are conferred mainly by mucins. Mucins are large heavily glycosylated proteins and typically have a molecular mass higher than 106 Da. Mucins, however, are predominantly decorated with O-glycan sugars, which accounts for up to 80% of their molecular weight. The diverse site-specific and mucin-specific glycosylation patterns influence the properties of the mucin and therefore the mucus gel. It is well known that mucin glycosylation is altered in infection and disease (Arike et al., 2017; Hasnain et al., 2017). Here we describe methods to assess the amounts of intestinal mucins in murine models and assess the changes in glycosylation with a particular focus on sialylation and sulphation of mucins. Previous methods have not differentiated between mucins isolated from the secreted barrier or those stored within the goblet cells. Methods described here can be employed to assess the changes in secreted or goblet cell-stored mucins in each individual animal. Moreover, using high-iron diamine staining changes in amounts of mucins can also be correlated this with changes in mucin glycosylation.
Materials and Reagents
- Pipette tips (Thermo Fisher Scientific, FinntipTM Pipette Tip)
- 19.5 gauge needle (BD, catalog number: 305187 )
- Needles (BD PrecisionGlideTM Needle)
- Nitrocellulose membrane (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 88018 )
- BIOMAXTM BML film (Carestream Health, catalog number: 876-1520 )
- Microscope slides (Menzel Gläser, SuperFrost® Plus) (VWR, catalog number: 631-9483 )
- Cover slips (Fisher Scientific, catalog number: 12-546-2 )
- Histology cassettes, moulded lid (ProSciTech, catalog number: RCH40-W )
- Biopsy Pads, 100% Reticulated Foam (Trajan Scientific and Medical, catalog number: BPBL )
- Polystyrene cube
- 10 cm Petri dishes (Corning, catalog number: 430591 )
- Transfer pipette, polyethylene (Sigma-Aldrich, catalog number: Z350826-500EA )
- Cell scraper (VWR, catalog number: 734-0386 )
- D-TubeTM dialyzers (Merck, UK)
- C57BL/6 mice (Animal Resource Centre, WA, Australia)
- 100% ethanol (Sigma-Aldrich, catalog number: 443611 )
- Non-sterile phosphate buffered saline (PBS) (Thermo Fisher Scientific, catalog number: 10010031 )
- Urea (VWR, BDH®, catalog number: BDH4602 )
- Guanidinium chloride (GuCl) (Sigma-Aldrich, catalog number: G3272 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884 )
- Dithiothrethiol (DTT) (Melford Laboratories, catalog number: MB1015 )
- Iodoacetamide (Sigma-Aldrich, catalog number: I6125 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- Bromophenol blue (Sigma-Aldrich, catalog number: B0126 )
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771 )
- Tris base (Roche Diagnostics, catalog number: 10708976001 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653 )
- Sodium citrate (Sigma-Aldrich, catalog number: PHR1416 )
- Periodic acid (Sigma-Aldrich, catalog number: P7875 )
- Acetic acid (Sigma-Aldrich, catalog number: A9967 )
- Sodium metabisulphite (Sigma-Aldrich, catalog number: 31448 )
- Schiff’s reagent (Sigma-Aldrich, catalog number: 3952016 )
- N,N-dimethyl-p-phenylenediamine (HCl) (Sigma-Aldrich, catalog number: D5004 )
- N,N-dimethyl-M-phenylenediamine (HCl)2 (Sigma-Aldrich, catalog number: 219223 )
- Iron(III) chloride (FeCl3) (Sigma-Aldrich, catalog number: 157740 )
- Alcian blue 8GX (AB) (Sigma-Aldrich, catalog number: A5268 )
- Tween-20 (Sigma-Aldrich, catalog number: P1379 )
- Skimmed milk powder (Sigma-Aldrich, catalog number: 1443825 )
- Odyssey® blocking buffer (LI-COR, catalog number: P/N 927-40000 )
- Muc2 antibody: Muc2.3, Rabbit polyclonal antibody (made in house) or H300 Muc2, Rabbit polyclonal antibody (Santa Cruz Biotechnology, catalog number: sc-15334 )
- Enhanced chemiluminescence substrate, Western Lightning® Plus-ECL (PerkinElmer, catalog number: NEL105001EA )
- IRDye® 800CW Donkey anti-Rabbit IgG (H+L) (LI-COR, catalog number: P/N 925-32213 )
- Peroxidase AffiniPure Donkey Anti-Rabbit IgG (H+L) (Jackson ImmunoResearch, catalog number: 711-035-152 )
- Alkaline Phosphatase AffiniPure Goat Anti-Rabbit IgG (H+L) (Jackson ImmunoResearch, catalog number: 111-055-003 )
- Haematoxylin (Sigma-Aldrich, catalog number: H3136 )
- Nitro Blue Tetrazolium (Tablet) (Sigma-Aldrich, catalog number: N5514 )
- Wax (MÜNZING, catalog number: CERETAN WE 3501 )
- Xylene (Sigma-Aldrich, catalog number: 214736 )
- Potassium hydroxide (KOH) (Sigma-Aldrich, catalog number: 484016 )
- Pertex mounting medium (MEDITE, catalog number: 41-4012-00 )
- 10% neutrally buffered formalin (diluted from CONFIX PURPLE, 10% NBF X5 concentrate) (Australian Biostain, catalog number: ACFP )
- Guanidinium chloride (GuCl) reduction buffer (see Recipes)
- Loading buffer (see Recipes)
- TAE buffer (see Recipes)
- 4x SSC buffer (see Recipes)
- Periodic acid solution (see Recipes)
- Sodium metabisulphite solution (see Recipes)
- High Iron Diamine solution (see Recipes)
- Alcian blue solution (see Recipes)
- TBST buffer (see Recipes)
- Milk blocking buffer (see Recipes)
Equipment
- Curved tweezers
- Pipettes (Eppendorf, model: Research® plus )
- Spring scissor (Fine Science Tools, catalog number: 15018-10 )
- Forceps (World Precision Instruments, catalog number: 501216 )
- 15 ml Falcon conical tube rotor (Beckman Coulter, model: C1015 )
- Incubator
- Electrophoresis gel tank (Figure 1) (Plaztek Scientific, catalog number: Wini-000 )
- Electrophoresis power pack (Figure 1) (Select Bioproducts, model: BioVolt 250V )
- Vacuum Blotting Unit with Bio-Rad Blotter (Figure 2) (Bio-Rad Laboratories, model: Model 785 )
- Odyssey® CLx Imager (LI-COR, model: Odyssey® CLx )
- Micro Spin tissue processor (STP) 120 (Microm UK Ltd.)
- Micron Cool Cut HM355S microtome (Microm, model: HM355S )
- Microtome blade MX35 (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 3052835 )
- Water bath (Thermoline Scientific, catalog number: TWB-D-SERIES )
- Olympus bright field microscope (Olympus Tokyo, Japan)
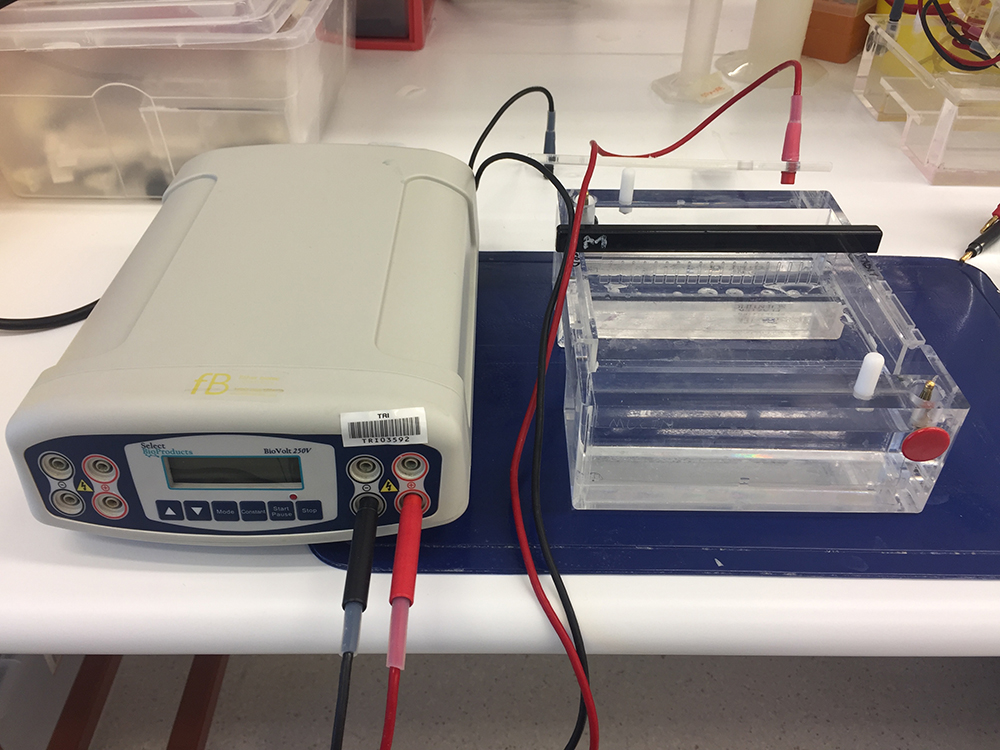
Figure 1. Electrophoresis gel tank (right) and electrophoresis power supply (left) for gel electrophoresis analyses of mucins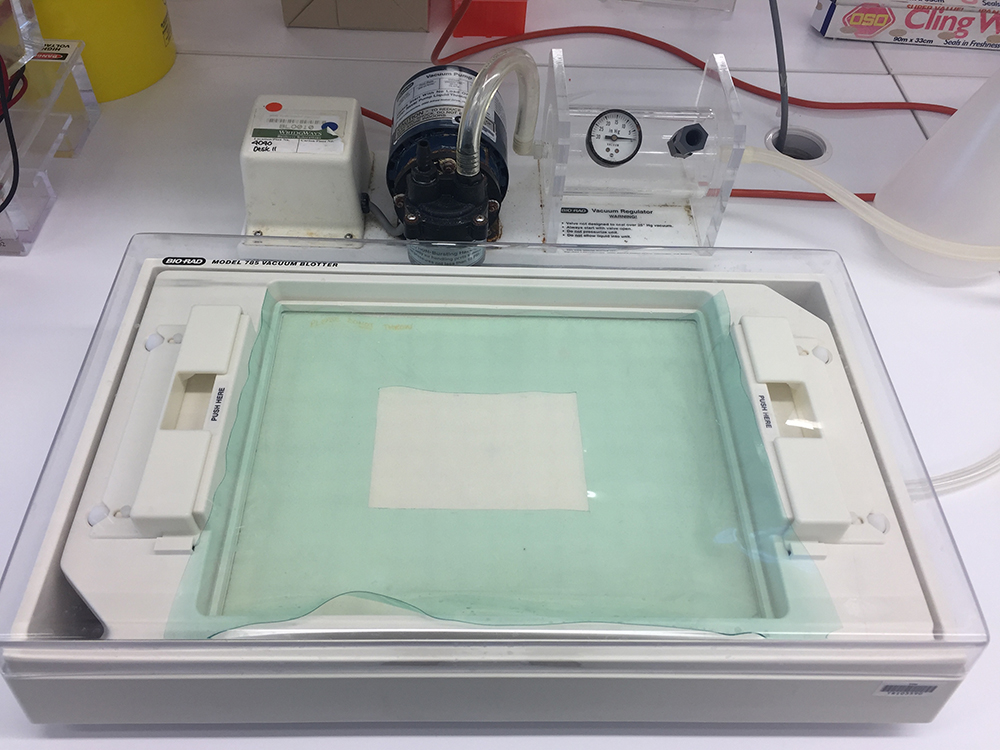
Figure 2. Vacuum Blotting Unit with vacuum blotter chamber (front) and vacuum pump (back)
Software
- NIS-Elements software v.3.0 (Nikon, Tokyo, Japan)
- Image StudioTM Lite software (LI-COR Biosciences)
Procedure
An outline is provided for all procedures included in this protocol (Figure 3)
Figure 3. Outlines of the procedures described
- Biochemical analysis of mucins and mucus
- Mice are euthanized using cervical dislocation according to approved ethical guidelines by the host institute in a PC2 laboratory.
- Mice are sprayed with 70% ethanol to avoid the contamination of samples with hair.
- A mid-line incision is made using scissor in the abdominal cavity.
- The intestine is pulled out gently using curved tweezers. Intestines are cut at the end attached to the stomach and the rectum end and dissected out. Small intestine, caecum and colon should be kept separately for analysis.
- Two different types of analyses can be conducted; total mucus present in the murine intestine to assess the overall changes in the mucus barrier or the secreted mucus which covers and protects the intestinal epithelium.
For secreted mucus extraction from mouse intestine- Place the intestine in a 10 cm Petri dish. Gently flush the intestines with PBS, to remove fecal matter, using a syringe with a 19.5 gauge needle through one open end. The amount of PBS will vary depending on the mouse strain and/or the part of intestine that are used. Usually up to 5 ml is used for colon, 7 ml for the small intestine and approximately 3-4 ml for the caecum. This material is discarded unless required for microbiota analyses.
- Place the intestine in a new Petri dish and flush the intestine slowly 5 times with 3 M urea (2 ml each flush) to obtain the secreted mucus from the lumen of the intestine. 3 M urea is recommended, as higher concentrations of urea or the use of GuCl would result in disrupting the intestinal epithelial cell layer. The material isolated is viscous; therefore use a plastic transfer pipette to transfer the material to a 15 ml Falcon tube on ice.
- Intestinal tissue is processed for histology (as described in Procedure B–histochemical analysis of mucin glycoproteins) before (a small section from the intestine) and after flushing (the whole intestine is rolled) with urea to ensure that only the secreted mucus was isolated and the intestinal crypts remained intact.
For total mucus extraction from mouse intestine- Place the intestine in a 10 cm Petri dish. Gently flush intestine with PBS, to remove fecal matter, using a syringe with a 19.5 gauge needle through one open end. It is important to ensure that the intestine is only gently flushed; otherwise the secreted mucus layer will be lost. The amount of PBS will vary depending on the strain of mice and/or the part of intestine that are used. Usually up to 5 ml is used for colon, 7 ml for the small intestine and approximately 3-4 ml for the caecum. This material is discarded as it consists of insoluble fecal matter and negligible amounts of secreted mucus (as has been previously investigated by the authors, the faecal mucin matter is not detected via Western blotting).
- Cut intestine open longitudinally using a spring scissor. Once open, scrape off the residual mucus, using a cell scraper. This will remove the mucins present in the goblet cells as well as the secreted mucus layer and represents the total intestinal mucus.
- The mucus is transferred to a 15 ml Falcon tube and solubilized in 6 M GuCl reduction buffer (see Recipes) at 4 °C overnight by rotation using a Beckman Coulter 15 ml rotator. GuCl is a strong chaotrope and denatures proteins and is recommended for the solubilisation of total mucus isolated from animals. Once solubilized, store sample at -80 °C until required later.
- Place the intestine in a 10 cm Petri dish. Gently flush the intestines with PBS, to remove fecal matter, using a syringe with a 19.5 gauge needle through one open end. The amount of PBS will vary depending on the mouse strain and/or the part of intestine that are used. Usually up to 5 ml is used for colon, 7 ml for the small intestine and approximately 3-4 ml for the caecum. This material is discarded unless required for microbiota analyses.
- Separation of mucins by agarose gel electrophoresis
- The total mucus and or secreted mucus can now be analysed using agarose gel electrophoresis.
- Before gel electrophoresis, reduce mucus samples with 50 mM DTT for 2 h at 37 °C to break up the disulphide bonds.
- After reduction, add 0.125 M iodoacetamide to the mucus sample and incubate for 30 min at room temperature in the dark to carboxymethylate mucus, to prevent reformation of the disulphide bonds.
- Up to 3 ml of the isolated samples in GuCl can then be dialyzed in 3-6 M urea using D-TubeTM dialyzers (Mw cut-off of 14 kDa or below is sufficient). Dialysis to urea is essential as GuCl has a very high salt content.
- 10% (v/v) loading buffer (see Recipes) is added in samples before electrophoresis.
- Samples are electrophoresed on 0.7-1% (w/v) agarose gels (around 5-7 mm in thickness) in TAE buffer (see Recipes) with 0.1% SDS at 30 V for 15 h (Thornton et al., 2000) (Figure 1).
- After electrophoresis, transfer mucins onto a nitrocellulose membrane by vacuum blotting in 4x SSC running buffer (see Recipes) at a pressure of 45-50 mbar using a 785 Bio-Rad Vacuum Blotter (Sheehan and Thornton, 2000) for around 1.5-3 h (Figure 2). To ensure a tight seal, 0.7% agarose can be used to seal the sides of the gel during the vacuum blotting process. Once the entire loading dye has disappeared from the gel, the vacuum blotting process is stopped and the gel is disposed. Note, vacuum blotting consistently results in efficient mucin protein transfer and is the recommended method. The nitrocellulose membrane is carefully removed using tweezers and can be analysed by using three distinct methods (as described in steps A7-A9).
To determine the changes in the glycosylation of mucins, two separate methods can be utilized which are described in detail below. Periodic Acid Schiff’s-Alcian blue staining can differentiate between acidic and neutral mucins, whereas High Iron Diamine-Alcian Blue staining can be used to identify the changes in mucin sulphation and sialylation.
- The total mucus and or secreted mucus can now be analysed using agarose gel electrophoresis.
- Glycoprotein detection using PAS staining
- Volumes of reagents used in this protocol are dependent on the size of the nitrocellulose membrane and should be sufficient to cover the membrane completely.
- Wash nitrocellulose membrane with dH2O.
- Incubate nitrocellulose membrane with periodic acid solution (see Recipes) at room temperature for 30 min. Periodic acid will oxidise the vicinal diols in mucin glycoproteins.
- Wash membrane thoroughly with dH2O to remove the periodic acid solution.
- Nitrocellulose membrane is incubated with sodium metabisulphite solution (see Recipes) for 5 min, the solution is discarded and this step is repeated once more. This step is essential to reduce background staining.
- Add Schiff’s reagent to the membrane and incubate for approximately 10-15 min at room temperature. The vicinal diols in mucin glycoproteins exposed by periodic acid will react with the Schiff’s reagent to give a purple-magenta colour.
- Rinse membrane with sodium metabisulphite solution, followed by a wash with dH2O to stop the reaction.
- Air-dry nitrocellulose membrane before taking scanning using Odyssey® CLx Imager and densitometry analyses (Thornton et al., 1994) (Figure 4).
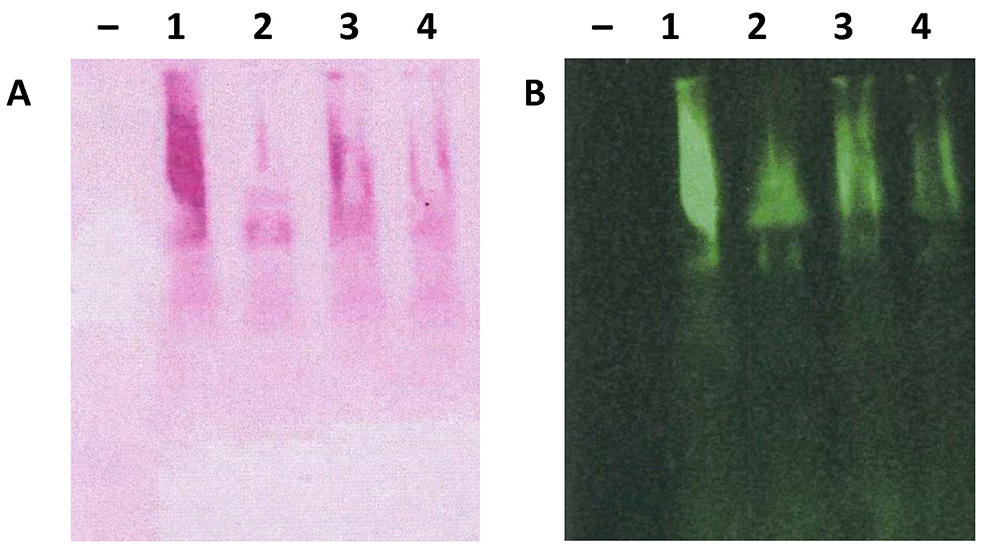
Figure 4. Scanned nitrocellulose membrane stained with PAS and anti-Muc2 antibody. Total mucus from the colon was extracted from 4 individual C57BL/6 mice and analysed using 1% agarose gel electrophoresis and transferred to nitrocellulose membrane. Nitrocellulose membrane was stained with PAS (A) or anti-Muc2 antibody (B, H300, from Santa Cruz) and developed using infrared-labelled secondary antibody. Membrane was scanned using the Odyssey® CLx Imager.
Note: Each individual animal colon yields sufficient mucus for gel electrophoresis analyses–indicates empty lane as negative control.
- Volumes of reagents used in this protocol are dependent on the size of the nitrocellulose membrane and should be sufficient to cover the membrane completely.
- Detection of sulphated glycoprotein using High Iron Diamine (HID)
- Volumes of reagents used in this protocol are dependent on the size of the nitrocellulose membrane and should be sufficient to cover the membrane completely.
- Wash nitrocellulose membrane with dH2O.
- Incubate nitrocellulose membranes with HID solution (see Recipes) for 18 h at room temperature.
- Wash membrane in dH2O and air-dry before scanning using Odyssey® CLx Imager (Hasnain et al., 2017).
- Volumes of reagents used in this protocol are dependent on the size of the nitrocellulose membrane and should be sufficient to cover the membrane completely.
- Immunodetection of mucin proteins
- Volumes of reagents used in this protocol are dependent on the size of the nitrocellulose membrane and should be sufficient to cover the membrane completely.
- Wash nitrocellulose membrane briefly in TBST buffer (see Recipes).
- Block membrane with milk blocking buffer (see Recipes) or Odyssey blocking buffer for at least 30 min at room temperature (alternatively membrane can be kept in blocking buffer at 4 °C overnight).
- Wash membrane 2 times with 15 min incubation for each wash using TBST buffer.
- Incubate with anti-Muc2 antibody (diluted in blocking buffer) overnight at 4 °C.
- Wash membrane 2 times with 5 min incubation for each wash using TBST buffer.
- Incubate with appropriate secondary antibody diluted in blocking buffer for 30 min at room temperature.
- For horseradish peroxidase (HRP)-conjugated secondary antibodies, peroxidase conjugated donkey anti-rabbit IgG is incubated with membrane at 1 in 10,000 dilution in TBST for 1 h at room temperature. Membrane is then washed 3 times with 5 min incubation for each wash using TBST buffer. Incubate membrane with enhanced chemiluminescence western detection reagent for approximately 2 min and then washed to remove excess and subsequently expose nitrocellulose membrane to BIOMAXTM BML film in the dark and develop.
- For the phosphatase conjugated secondary antibody, phosphatase conjugated goat anti-rabbit IgG is incubated with membrane at 1 in 10,000 dilution in TBST for 1 h at room temperature. After 3 times washes in TBST buffer, membrane is then incubated with nitro-blue tetrazolium (NBT)/5-bromo-4-chloroindol-3yl phosphate substrate for 5 min to develop.
- For infrared fluorescent dyes labelled antibodies, IRDye® 800CW donkey anti-rabbit IgG is incubated with membrane at 1 in 2,000 dilution in TBST for 1 h at room temperature. After 3 times washes in TBST buffer, nitrocellulose membranes can be directly scanned at 700 or 800 nm using Odyssey® CLx Imager (Figure 4B).
- Volumes of reagents used in this protocol are dependent on the size of the nitrocellulose membrane and should be sufficient to cover the membrane completely.
- Mice are euthanized using cervical dislocation according to approved ethical guidelines by the host institute in a PC2 laboratory.
- Histochemical analysis of mucin glycoproteins
- ‘Swiss Roll’ technique
- Dissect the intestine as described in steps A1-A4 (Figure 5A). Intestine is then cut open longitudinally (Figure 5B) using spring scissors and fecal matter is gently removed by using a P200 pipette tip (Figure 5C). Roll the intestine around a 16 gauge needle with the help of tweezers starting from the rectum end (Figures 5D and 5E). The roll is then pinned onto a small piece of polystyrene using 30 gauge needles (Figure 5F) and placed in fixative solution (4% PFA or 10% neutrally buffered formalin Figure 5G) for 24 h (Reilly and Kirsner, 1965).
- Dissect the intestine as described in steps A1-A4 (Figure 5A). Intestine is then cut open longitudinally (Figure 5B) using spring scissors and fecal matter is gently removed by using a P200 pipette tip (Figure 5C). Roll the intestine around a 16 gauge needle with the help of tweezers starting from the rectum end (Figures 5D and 5E). The roll is then pinned onto a small piece of polystyrene using 30 gauge needles (Figure 5F) and placed in fixative solution (4% PFA or 10% neutrally buffered formalin Figure 5G) for 24 h (Reilly and Kirsner, 1965).
- Tissue processing
- Swiss rolls are transferred to embedding cassettes (Figure 5H) and placed in 70% ethanol.
- Process specimens using the Micro Spin Tissue processor (STP) 120. Specimens then go through series of solvents (1 change of 70%, 90%, 95% ethanol, 3 changes of 100% ethanol, 3 changes of xylene and 3 changes of paraffin) for a total of 8 h before being embedded in wax.
- Paraffin cell blocks (Figure 5I) can be stored at room temperature until cut to a thickness of 4 µm sample using Micron Cool Cut HM355S microtome with microtome blade MX35.
- Sections are floated in a warm water bath (37.4 °C) before being positioned on microscope slides, dried at 37 °C overnight and stored at room temperature until use.
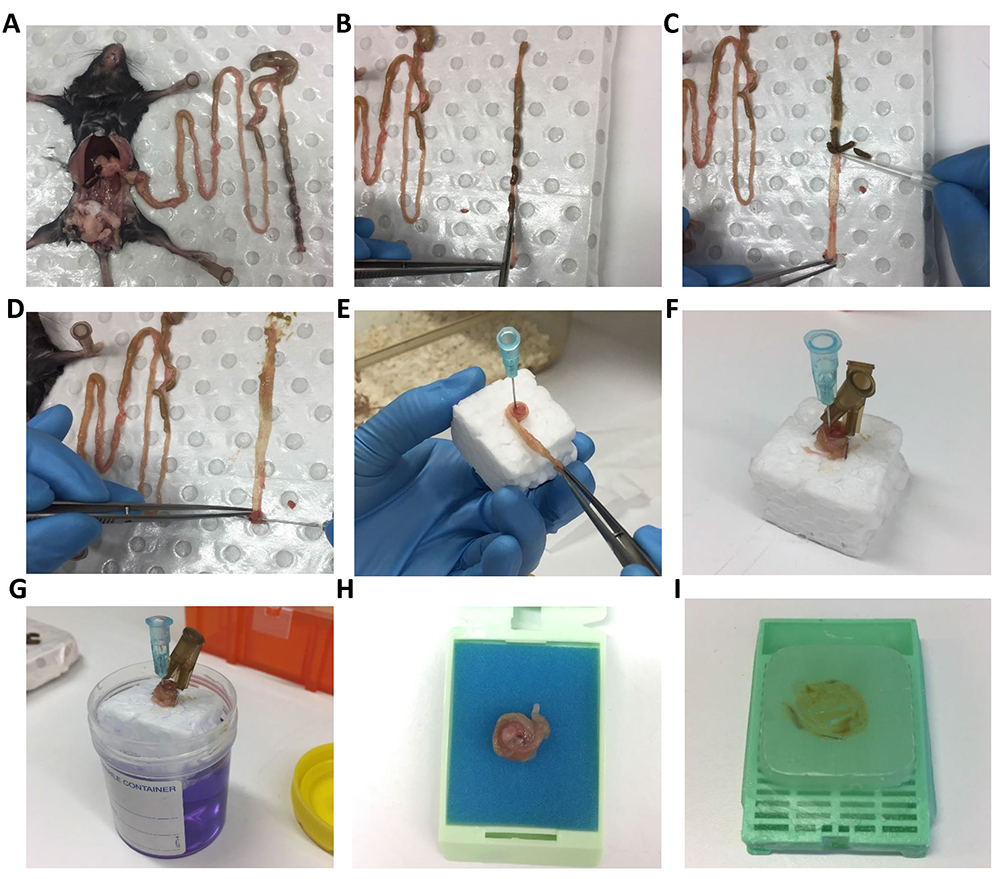
Figure 5. Mouse intestine dissection and ‘Swiss Roll’ technique for histology. Mouse intestine is dissected (A) followed by opening longitudinally using spring scissor (B) and removing feces using a P200 tip (C). Intestine is rolled around a 16 gauge needle with the help of tweezers (D, E). Swiss roll is secured on polystyrene cube with two needles pin in opposite direction (make sure needle is not placed in tissue) (F) before putting into specimen jar with 10% buffer formalin for fixation (G). Fixed Swiss roll is transferred into histology cassette (H) for paraffin embedding (I).
- Swiss rolls are transferred to embedding cassettes (Figure 5H) and placed in 70% ethanol.
- Periodic Acid Schiff’s-Alcian Blue (PAS-AB) staining
- Before staining, slides are rehydrated by going through xylene and ethanol gradient for 5 min in each solution (2 changes of xylene, 2 changes of 100% ethanol, 1 change of 95% and 70% ethanol). Rehydrated slides are placed in PBS for further staining.
- Place slides in AB solution for 5 min and then wash briefly in ddH2O. AB stains sulphated and sialylated glycoproteins.
- For staining of neutral glycoproteins, incubate the slides with 0.1 M potassium hydroxide in dH2O for 30 min at room temperature.
- Place slides in 1% periodic acid solution (see Recipes) for 5 min followed by a thorough wash in ddH2O to remove the acidic solution.
- The slides are then placed in Schiff’s reagent for 10 min and washed again in ddH2O.
- All slides are washed under running dH2O for 10 min and counterstained with haematoxylin for 30 sec.
- Slides are washed in hot ddH2O (warm ddH2O at high power, 30 sec in microwave) for 15 to 30 sec.
- Slides are placed in ethanol gradients (from 70% to 100%) and xylene for dehydration, mounted using Pertex mounting medium and sealed with cover slips (Figure 6).
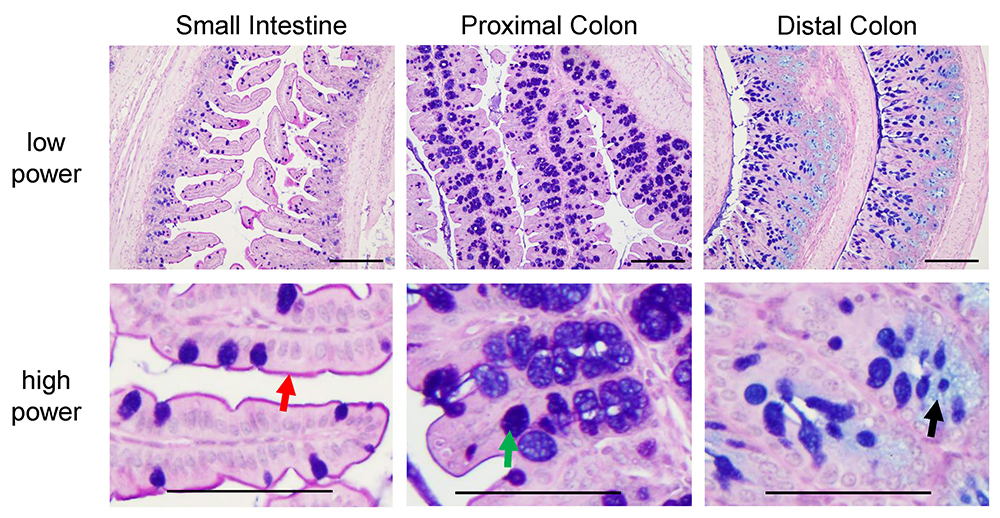
Figure 6. Pictures of PAS-AB stain in small intestine, proximal colon and distal colon of C57BL/6 mice. Neutral glycoproteins are stained in magenta (red arrow, depicting the glycocalyx), mucins that are both acidic and neutrally charged are stained in purple (blue plus magenta stain, green arrow) and acidic glycoproteins are stained in blue (black arrow). These slides were counterstained with haematoxylin (as described in step B3f). Scale bars = 200 µm (low power), 100 µm (high power).
- Before staining, slides are rehydrated by going through xylene and ethanol gradient for 5 min in each solution (2 changes of xylene, 2 changes of 100% ethanol, 1 change of 95% and 70% ethanol). Rehydrated slides are placed in PBS for further staining.
- High Iron Diamine-Alcian blue (HID-AB) staining
- Prepare HID solution freshly every time within 4-6 h of use (as described in Recipes).
- After the slides are passed through the xylene and ethanol gradient as described in step B3a, slides are incubated with HID for 18 h at room temperature. HID will stain sulphated mucins in black.
- Wash slides with ddH2O thoroughly to remove the HID staining solution.
- Place slides in AB solution for 30 min (Figure 7) (Spicer, 1965), which will stain sialylated mucins.
- No counterstaining is required; therefore slides are mounted as above (described in step B3h).
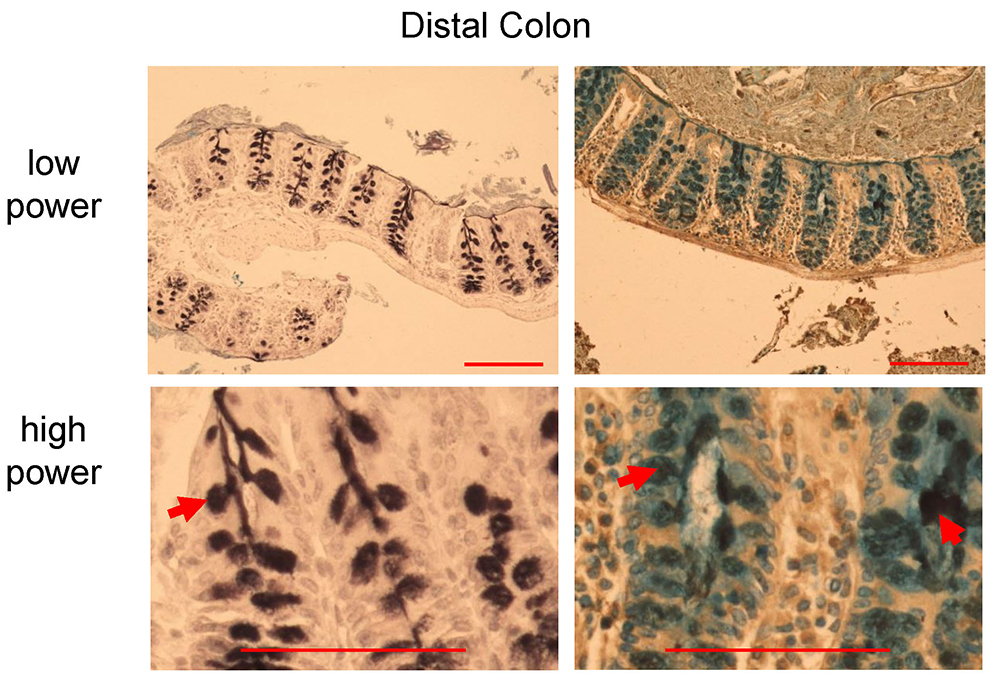
Figure 7. HID-AB staining in distal colon of C57BL/6 mice. Black stain indicates sulfomucins dominated area (left panel, red arrow), whereas goblet cells with sialomucins (blue) and mixed sulfo-sialomucins (black and blue colors in the right panel, red arrows). Scale bars = 200 µm (low power), 100 µm (high power).
- ‘Swiss Roll’ technique
Data analysis
- Densitometry analysis for mucin gel
- 2-3 biological samples should be included for quantification from each group.
- To quantify the band intensity of mucin gel, Image StudioTM Lite software is used. Multiple regions of interest with the same size are drawn over each mucin bands using the object drawing tool in the software. Intensity of bands is automatically calculated by software as arbitrary number. The number can be used for further comparison between groups.
- Quantification of histological staining
- Quantification is conducted on blinded samples.
- To quantify positive staining in tissue sections after histological staining, multiple pictures (usually 3-4 pictures) from one mouse should be taken. Pictures are taken randomly in areas with longitudinally sectioned crypts using an Olympus bright field microscope with a 40x lens in order to keep sampling process unbiased (although pictures can be taken with any lens, it is essential to keep the sampling procedure consistent). It is important to ensure longitudinally sectioned crypts are quantified, as it limits the variation, artifacts and the possibility of false negative staining.
- In each image, positive staining is defined by RGB pixel picking tool in NIS-Elements software v.3.0. Clicking the positive stained area using RGB pixel picking tool enables the software to pick areas that are positive for the exact same staining. This is achieved by defining the actual RGB pixel of the positive stained area. Background staining usually with a slightly lighter color (different pixel) will not be picked up by the software. The percentage of the longitudinally sectioned crypt area occupied by positive staining is calculated (Figure 8) for multiple crypts within each specimen and the average for that specimen determined, and used for statistical analysis.
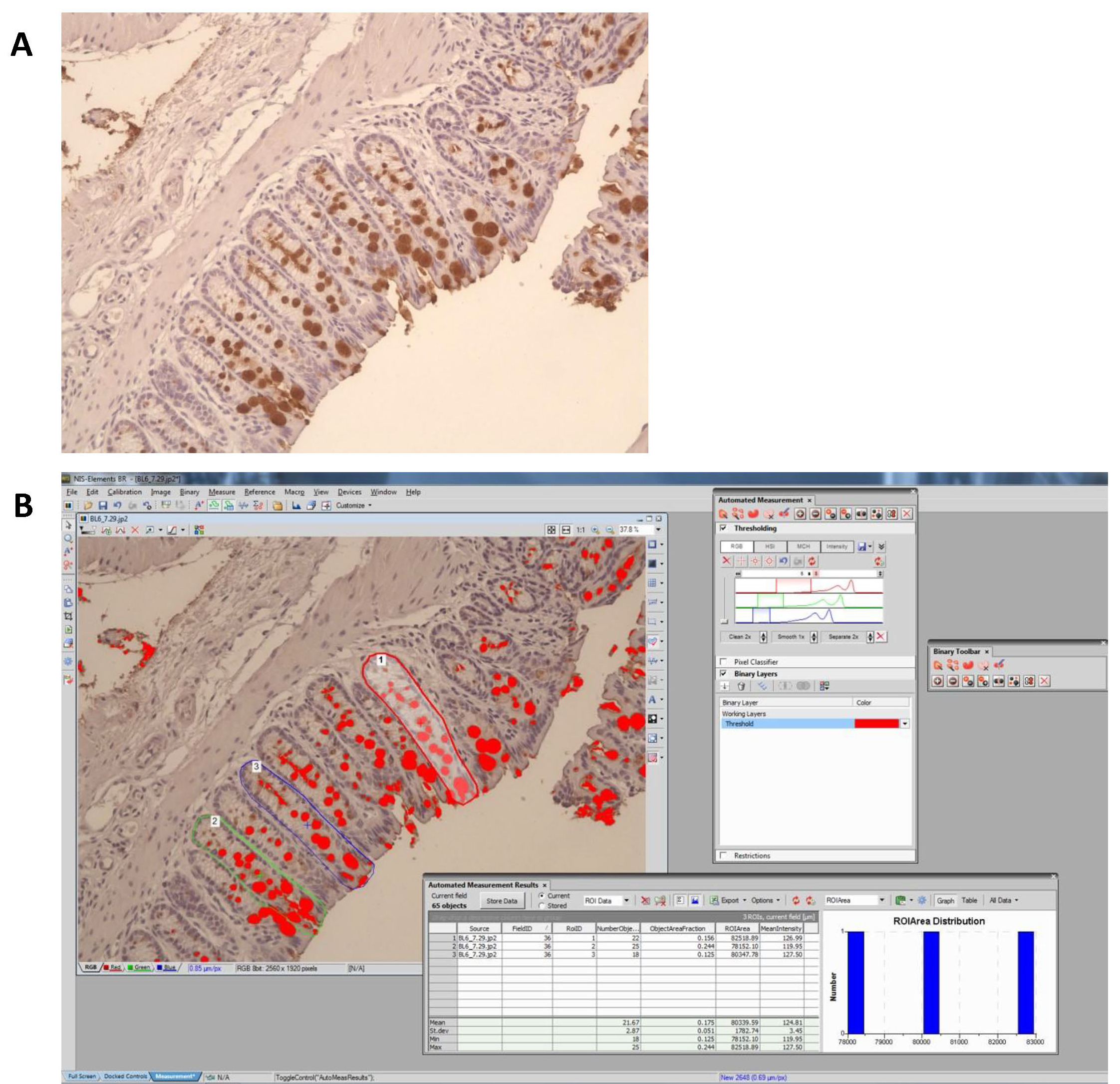
Figure 8. Quantification of positive staining by using NIS-Elements software. A. Original picture showing Muc2 positive staining in dark brown color. B. Positive staining was defined by RGB pixel picking tool in NIS-Elements software. For example, to quantify goblet cell volume in a longitudinally sectioned crypt, the percent of positive staining per crypt is calculated as the area covered by positive staining divided by the area of the crypt. Three to five individual crypts are analysed for each sample before taking the average value.
Recipes
- Guanidinium chloride (GuCl) reduction buffer
6 M GuCl dissolved in 0.1 M Tris-HCl buffer with 5 mM EDTA, pH 8.0 - Loading buffer
50% (v/v) glycerol in 10x TAE buffer
Bromophenol blue
1% (w/v) SDS - TAE buffer
40 mM Tris acetate in 1 mM EDTA solution, pH 8.0 - 4x SSC buffer
0.6 M sodium chloride and 60 mM sodium citrate in H2O, pH 7.0 - Periodic acid solution
1% (w/v) periodic acid in 3% (v/v) acetic acid solution - Sodium metabisulphite solution
0.1% (w/v) sodium metabisulphite in 0.01 M HCl solution - High Iron Diamine solution
120 mg of N,N-dimethyl-M-phenylenediamine (HCl)2
20 mg of N,N-dimethyl-p-phenylenediamine (HCl)
Dissolve in 50 ml of ddH2O
1.4 ml of 10% FeCl3 added immediately (prepare freshly every time) - Alcian blue solution
1 g of Alcian blue 8GX power
100 ml of 3% glacial acetic acid
Adjust pH to 2.5 using acetic acid - TBST buffer
150 mM NaCl in10 mM Tris-HCl pH 8.0 with 0.05% Tween-20
10.Milk blocking buffer
1% (w/v) skimmed milk powder in TBST buffer
Acknowledgments
A University of Queensland Postdoctoral Fellowship currently supports Sumaira Hasnain. The University Of Queensland Foundation Of Research Excellence Award to Sumaira Hasnain supports Ran Wang. The diamine method was originally described by Spicer (Spicer, 1965) and has been modified for our studies. David Thornton, John Sheehan and Ingemar Carlstedt (Thornton et al., 1994) originally developed the method of assessing mucins in human samples using western blotting on nitrocellulose membrane which was adapted for the animal studies.
References
- Arike, L., Holmen-Larsson, J. and Hansson, G. C. (2017). Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology 27(4): 318-328.
- Hasnain, S. Z., Dawson, P. A., Lourie, R., Hutson, P., Tong, H., Grencis, R. K., McGuckin, M. A. and Thornton, D. J. (2017). Immune-driven alterations in mucin sulphation is an important mediator of Trichuris muris helminth expulsion. PLoS Pathog 13(2): e1006218.
- Reilly, R. W. and Kirsner, J. B. (1965). Runt intestinal disease. Lab Invest 14: 102-107.
- Sheehan, J. K. and Thornton, D. J. (2000). Heterogeneity and size distribution of gel-forming mucins. Methods Mol Biol 125: 87-96. Spicer, S. S. (1965). Diamine methods for differentialing mucosubstances histochemically. J Histochem Cytochem 13: 211-234.
- Spicer, S. S. (1965). Diamine methods for differentialing mucosubstances histochemically. J Histochem Cytochem 13: 211-234.
- Thornton, D. J., Carlstedt, I. and Sheehan, J. K. (1994). Identification of glycoproteins on nitrocellulose membranes and gels. Methods Mol Biol 32: 119-128.
- Thornton, D. J., Khan, N. and Sheehan, J. K. (2000). Separation and identification of mucins and their glycoforms. Methods Mol Biol 125: 77-85.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wang, R. and Hasnain, S. Z. (2017). Analyzing the Properties of Murine Intestinal Mucins by Electrophoresis and Histology. Bio-protocol 7(14): e2394. DOI: 10.21769/BioProtoc.2394.
Category
Immunology > Mucosal immunology > Digestive tract
Cancer Biology > General technique > Biochemical assays > Protein analysis
Biochemistry > Protein > Electrophoresis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










