- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fluorophore Labeling, Nanodisc Reconstitution and Single-molecule Observation of a G Protein-coupled Receptor
Published: Vol 7, Iss 12, Jun 20, 2017 DOI: 10.21769/BioProtoc.2332 Views: 10232
Reviewed by: Arsalan DaudiKenji SugiyamaStéphane Roméro

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

FRET-based Microscopy Assay to Measure Activity of Membrane Amino Acid Transporters with Single-transporter Resolution
Didar Ciftci [...] Olga Boudker
Apr 5, 2021 6772 Views

Single-Molecule Studies of Membrane Receptors from Brain Region Specific Nanovesicles
Surya P. Aryal [...] Christopher I. Richards
May 20, 2021 5966 Views

Single Protein Detection and Imaging with Evanescent Scattering Microscopy
Pengfei Zhang [...] Shaopeng Wang
Oct 20, 2022 1892 Views
Abstract
Activation of G protein-coupled receptors (GPCRs) by agonist ligands is mediated by a transition from an inactive to active receptor conformation. We describe a novel single-molecule assay that monitors activation-linked conformational transitions in individual GPCR molecules in real-time. The receptor is site-specifically labeled with a Cy3 fluorescence probe at the end of trans-membrane helix 6 and reconstituted in phospholipid nanodiscs tethered to a microscope slide. Individual receptor molecules are then monitored over time by single-molecule total internal reflection fluorescence microscopy, revealing spontaneous transitions between inactive and active-like conformations. The assay provides information on the equilibrium distribution of inactive and active receptor conformations and the rate constants for conformational exchange. The experiments can be performed in the absence of ligands, revealing the spontaneous conformational transitions responsible for basal signaling activity, or in the presence of agonist or inverse agonist ligands, revealing how the ligands alter the dynamics of the receptor to either stimulate or repress signaling activity. The resulting mechanistic information is useful for the design of improved GPCR-targeting drugs. The single-molecule assay is described in the context of the β2 adrenergic receptor, but can be extended to a variety of GPCRs.
Keywords: G-protein coupled receptorsBackground
GPCRs mediate cellular communications, both locally and over long distances, especially in the endocrine system. For instance, the cellular response to hormones such as adrenaline is mediated through adrenergic receptors, of which the β2 adrenergic receptor (β2AR) is a prominent member. β2AR is expressed throughout the human body and is especially important in pulmonary, cardiac and immunological systems. Pharmacologically, agonists targeting β2AR are medically proven to alleviate acute asthma attacks, since activation of β2AR relaxes smooth muscle lining in the respiratory tracts. At the molecular level, β2AR binds extracellular ligands and transmits signals across the cell membrane to intracellular effectors, such as G proteins or β arrestin. A variety of β2AR ligands are known, and these are classified as agonists or inverse agonists, depending on whether they stimulate signaling activity or reduce the activity below the basal level, respectively (Baker, 2010). Crystallographic studies have revealed the three-dimensional structures of β2AR in both inactive (Cherezov et al., 2007) and active (Rasmussen et al., 2011) conformations. However, less is known about how the receptor converts from the inactive to active conformation during signaling and how these transitions are linked to ligand binding.
Observation of single receptor molecules can reveal spontaneous conformational transitions and the influence of ligands on conformational switching, providing a unique perspective on receptor activation (Lamichhane et al., 2015). Development of a single-molecule assay requires labeling the receptor with a fluorescent reporter group at an informative position and a method for reconstituting individual receptor molecules in a membrane-like environment. A Cy3 fluorophore attached to the cytoplasmic end of trans-membrane helix 6 is a suitable reporter of conformational transitions, since the fluorescence quantum yield is sensitive to the local protein environment, a phenomenon referred to as protein-induced fluorescence enhancement (Hwang et al., 2011; Stennett et al., 2015). Moreover, phospholipid nanodiscs provide an ideal system for reconstitution and observation of single receptor molecules (Bayburt and Sligar, 2010). Individual labeled receptors in nanodiscs can be monitored over extended time periods by total internal reflection fluorescence (TIRF) microscopy, directly revealing transitions between inactive and active-like receptor conformations. The concept of the assay is illustrated in Figure 1. Statistical analysis of a collection of receptor molecules provides the rate constants for conformational exchange and the equilibrium distribution of the two conformational states, information that is difficult to obtain otherwise. These analyses can be readily performed in the absence or presence of various β2AR ligands, providing detailed mechanistic information on the linkage of receptor activation to ligand binding. Such information should be useful in the design of improved GPCR-targeting drugs with finely tuned pharmacological efficacies and reduced side effects. Here we describe the protocols to label β2AR with a Cy3 fluorophore, to reconstitute the labeled receptor in nanodiscs, to attach receptor-nanodisc complexes to a microscope slide and the procedures used to record and analyze single-molecule TIRF microscopy data. Although we describe these methods in the context of β2AR, they are equally applicable to a variety of GPCRs.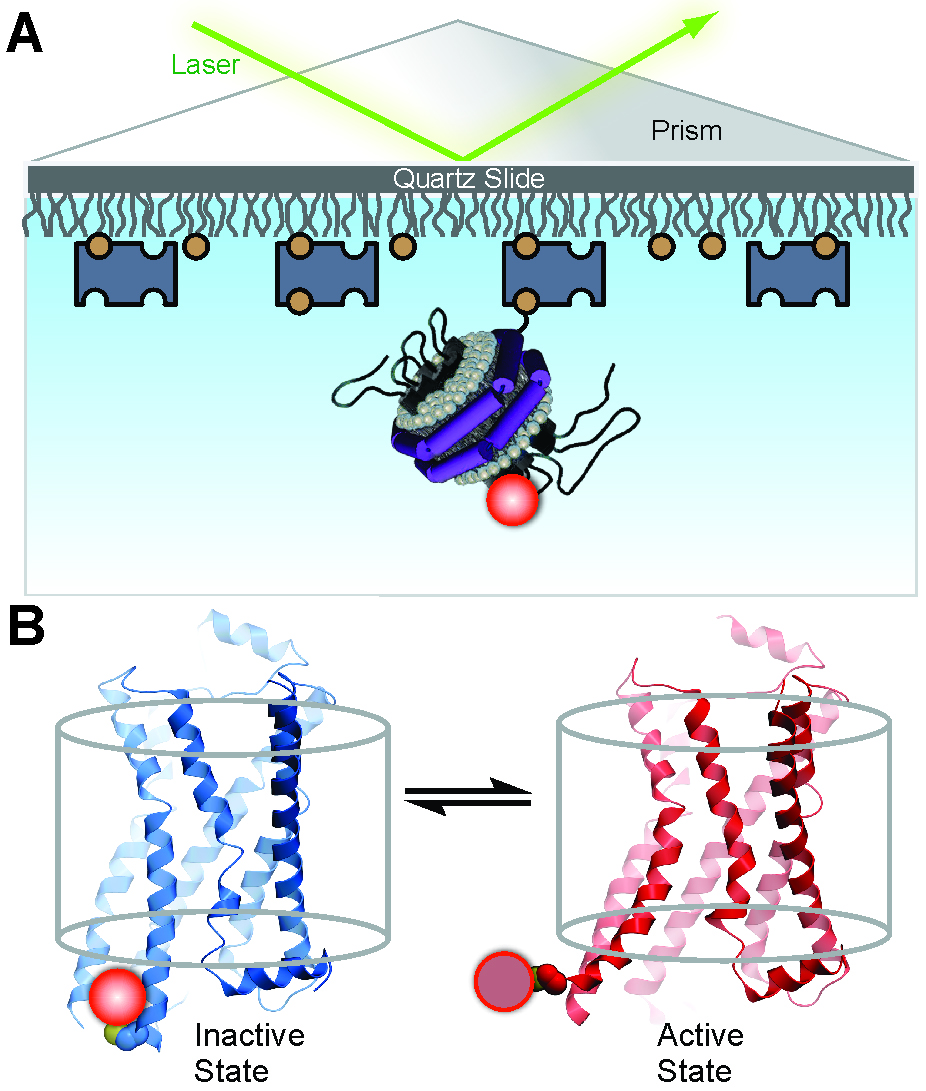
Figure 1. Experimental system to monitor conformational transitions of β2AR at the single-molecule level (reproduced from Lamichhane et al., 2015). A. An individual receptor molecule (black) labeled with Cy3 (red sphere) incorporated in a phospholipid nanodisc is tethered to a quartz surface coated with polyethylene glycol (wavy lines) via biotin (orange circles) and streptavidin (dark blue rectangles). The labeled receptor is illuminated in the evanescent field of a totally internally reflected 532 nm laser beam (green). The cartoon of the receptor-nanodisc complex is adapted from Bayburt and Sligar (2010). B. Expanded view of a single receptor-nanodisc complex, showing the receptor exchanging between inactive (blue) and active (red) conformations, with corresponding changes in the local environment of the Cy3 probe attached to Cys265 (light or dark red spheres, respectively). The transparent cylinder represents an abstraction of the lipid bilayer.
Materials and Reagents
- PD-10 desalting column (GE Healthcare, catalog number: 17085101 )
- 2 ml Eppendorf tube
- Quartz microscope slides 1” x 3” x 1 mm thick, with a small diameter hole drilled at each end (http://finkenbeiner.com/quartzslides.php)
- Microscope cover slips 22 x 40-1 (Fisher Scientific, catalog number: 12-545-C )
- Double sided tape (Scotch, 3M)
- Pipette tip
- 100 kDa MWCO vivaspin 2 concentrators polyethersulfon (PES) membrane (Sartorius, catalog number: VS1041 )
- Cy3 maleimide, mono-reactive dye (GE Healthcare, catalog number: PA13131 )
- Timolol maleate salt (Sigma-Aldrich, catalog number: T6394 )
- cOmpleteTM, Mini, EDTA-free protease inhibitor cocktail (Roche Diagnostics)
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888 )
- Talon metal affinity resin (Takara Bio, catalog number: 635502 )
- Imidazole (Sigma-Aldrich, catalog number: I5513 )
- DMSO (Thermo Fisher Scientific, InvitrogenTM, catalog number: D12345 )
- SDS-PAGE gels (Invitrogen NuPAGE Bis-Tris Pre-cast gels)
- Purified membrane scaffold protein 1 (MSP1), expressed in E. coli, as described (Ritchie et al., 2009)
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) (Avanti Lipids Polar, catalog number: 850457P )
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) (Avanti Lipids Polar, catalog number: 840034P )
- 16:0 biotinyl Cap PE (Avanti Lipids Polar, catalog number: 870277P )
- Bio-beads SM-2 resin (Bio-Rad Laboratories, catalog number: 1523920 )
- Ni-NTA resin (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 88222 )
- Glycerol (Sigma-Aldrich, catalog number: G6279 )
- Grease (Borer Chemie, catalog number: glisseal N )
- Neutravidin (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 31000 )
- 4-(2-hydroyethyl)-1-piperazineethanesulfonic acid (HEPES) (Sigma-Aldrich, catalog number: H3375 )
- Magnesium chloride (MgCl2) (anhydrous) (Sigma-Aldrich, catalog number: M8266 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 )
- Ethylenediaminetetraacetic acid (EDTA) (0.5 M solution) (Sigma-Aldrich, catalog number: 03690 )
- N-dodecyl- β-D-maltopyranoside (DDM) (Anatrace, catalog number: D310 )
- Cholesteryl hemisuccinate (CHS) (Sigma-Aldrich, catalog number: C6512 )
- ATP (Sigma-Aldrich, catalog number: A26209 )
- Phosphate-buffered saline (PBS) (Fisher Scientific, catalog number: 70-011-044 )
- Trolox (Acros Organics, catalog number: 218940050 )
- Glucose (Sigma-Aldrich, catalog number: 158968 )
- Glucose oxidase (Sigma-Aldrich, catalog number: G2133 )
- Catalase (Sigma-Aldrich, catalog number: C3155 )
- Low salt wash buffer (see Recipe)
- High salt wash buffer (see Recipe)
- Solubilization buffer (see Recipe)
- Wash buffer 1 (see Recipe)
- Wash buffer 2 (see Recipe)
- Labeling buffer (see Recipe)
- SEC elution buffer (see Recipe)
- IMAC elution buffer (see Recipe)
- Final buffer (see Recipe)
- Imaging buffer (see Recipe)
Equipment
- 100 ml glass tissue homogenizer (Sigma-Aldrich, catalog number: T2567 )
- Beckman Ultra centrifuge
- Ti45 rotor (Beckman Coulter, model: Type 45 Ti , catalog number: 339160)
- Ti70 rotor (Beckman Coulter, model: Type 70 Ti , catalog number: 337922)
- Beckman X-12R centrifuge(Beckman Coulter, model: Allegra® X-12R ) or equivalent
- AKTAxpress FPLC system (GE Healthcare, model: AKTAxpress )
- Superdex 200 Increase 100/300 size exclusion column (GE Healthcare, catalog number: 17517501 )
- Chromatography column packed with 1-2 ml Ni-NTA agarose beads (referred to below as Ni column)
- Axiovert 200 microscope (Carl Zeiss, model: Axiovert 200 ) or equivalent
- Water-immersion C-Apochromat 63x/1.2 W objective (Carl Zeiss, model: C-Apochromat 63x/1.2 W Corr ) or equivalent
- Charge-coupled device (EMCCD) camera (Andor Technology, model: DU-897E iXon+ EMCCD ) or equivalent
- 532 nm (green) laser (CrystaLaser, catalog number: CL532-050-S ) or equivalent
Software
- Data acquisition software (available from https://cplc.illinois.edu/software/)
- MATLAB scripts
- Igor software
Procedure
The protocols described here were developed for a beta 2 adrenergic receptor (β2AR) construct containing a thermo-stabilizing E122W mutation (Roth et al., 2008), C-terminal truncation at residue 348, deletion of residues 245 to 259 in the third intracellular loop (ICL3), a FLAG tag at the N-terminus and a 10x His tag at the C-terminus. The construct also contains C327S and C341S mutations, leaving just a single reactive cysteine residue (Cys265) for fluorophore labeling. The deletions, truncations and cysteine mutations do not disrupt binding of ligands to the receptor (Liu et al., 2012). The procedures to express the receptor in sf9 insect cells infected with high-titer recombinant virus are described elsewhere (Cherezov et al., 2008).
- Receptor purification and Cy3 labeling
- Lyse and wash frozen β2AR biomass (5 L, roughly 0.5 mg receptor/1 L biomass) using a 100 ml glass tissue homogenizer. Cell lysis is chiefly achieved through the use of hypertonic (low salt) buffer and the mechanical force of homogenization. Wash lysed biomass with the low salt wash buffer and the high salt wash buffer 3 times, respectively (500 ml per wash). Use Beckman Ultra centrifuge to pellet membrane fragment suspension (158,000 x g, Ti45 rotor, 35 min, 4 °C) in between washes. The washed membranes can be stored at -80 °C for up to two weeks or 4 °C overnight. Ideally, the washed membrane should be aliquoted, so that each aliquot of membranes contains roughly 0.5-1 mg of final purified receptor.
- Treat one aliquot of washed membrane with 1 mM timolol (dissolved in ddH2O), 1 crushed Roche protease inhibitor tablet for 30 min at 4 °C. This step can be done by adding a previously prepared 100 mM timolol stock (stored at 4 °C) and crushed Roche inhibitor directly to the suspended membrane. Then, thoroughly mix the treated membrane with the solubilization buffer at 1:1 ratio for 3 h at 4 °C.
- Remove membrane pellet from supernatant using Beckman Ultra centrifuge (rotor Ti70) at 265,000 x g, 35 min, 4 °C. Add stock 5 M NaCl, 4 M imidazole to the supernatant, so that the final concentration is 800 mM NaCl, 20 mM imidazole. Add 0.5 ml washed talon slurry to the supernatant and incubate at 4 °C overnight.
- Pellet talon resin using Beckman X-12R centrifuge (or equivalent) at 500 x g, 1 min, 4 °C and remove supernatant. Wash with 50 ml wash buffer 1, 10 ml wash buffer 2, 10 ml labeling buffer. Add 5 ml labeling buffer on top of the pelleted resin, and 20 μl of 5 mg/ml (in DMSO) Cy3 stock (approximately 25 μM final concentration, whereas the receptor concentration is approximately 1-5 μM). Incubate at 4 °C for 2 h in the dark.
- Load talon resin onto an empty column and wash with 50 ml wash buffer 2 using gravity flow. Elute purified, labeled receptor using 1 ml fractions of elution buffer. Run SDS-PAGE to visualize the elution pattern. Concentrate Cy3-labeled receptors using a 100 kDa MWCO vivaspin2 concentrator.
- Lyse and wash frozen β2AR biomass (5 L, roughly 0.5 mg receptor/1 L biomass) using a 100 ml glass tissue homogenizer. Cell lysis is chiefly achieved through the use of hypertonic (low salt) buffer and the mechanical force of homogenization. Wash lysed biomass with the low salt wash buffer and the high salt wash buffer 3 times, respectively (500 ml per wash). Use Beckman Ultra centrifuge to pellet membrane fragment suspension (158,000 x g, Ti45 rotor, 35 min, 4 °C) in between washes. The washed membranes can be stored at -80 °C for up to two weeks or 4 °C overnight. Ideally, the washed membrane should be aliquoted, so that each aliquot of membranes contains roughly 0.5-1 mg of final purified receptor.
- Nanodisc preparation
- Mix labeled receptor, purified MSP1 and phospholipid mixture at a molar ratio 1:10:700 in a 2 ml Eppendorf tube. The phospholipid mixture contains POPC, POPS and biotinyl Cap PE in the ratio 67.5%, 27.5%, and 5%. Add Bio-Bead resin and incubate overnight at 4 °C.
- Remove Bio-Bead resin by pelleting. Purify nanodiscs by size exclusion chromatography using an AKTAxpress FPLC system, superdex 200 Increase 100/300 GL column and SEC elution buffer. Representative elution profiles of membrane protein-nanodisc complexes are presented elsewhere (Ritchie et al., 2009). Remove empty nanodiscs by immobilized metal affinity chromatography (IMAC) using an AKTAxpress FPLC system, Ni column and IMAC elution buffer. Exchange buffer to the final buffer using a PD-10 desalting column (GE Healthcare).
Note: Ni-NTA and talon (Co2+) resins are both used to purify the receptor protein with poly-histidine tag, but differ in that 1) Ni2+ has greater affinity for histidine, conferring a larger binding capacity per ml of resin, whereas 2) Co2+ has greater selectivity toward histidine. As such talon resins are preferable to be used on crude lysate, as described in step A3 above, whereas Ni-NTA is preferable for the final purification step described here.
- Mix labeled receptor, purified MSP1 and phospholipid mixture at a molar ratio 1:10:700 in a 2 ml Eppendorf tube. The phospholipid mixture contains POPC, POPS and biotinyl Cap PE in the ratio 67.5%, 27.5%, and 5%. Add Bio-Bead resin and incubate overnight at 4 °C.
- Surface immobilization of receptor-nanodisc complexes
- Clean quartz slides and cover slips, passivate the surfaces with polyethylene glycol (5% biotinylated), as described previously (Lamichhane et al., 2010).
- Prepare a microfluidic channel about 4-5 mm wide on the quartz slide using double sided tape, fix cover slip on top of the double sided tape and close the two ends of channel using silicon grease at the time of experiment (Figure 2). This device is subsequently referred to as the sample chamber.
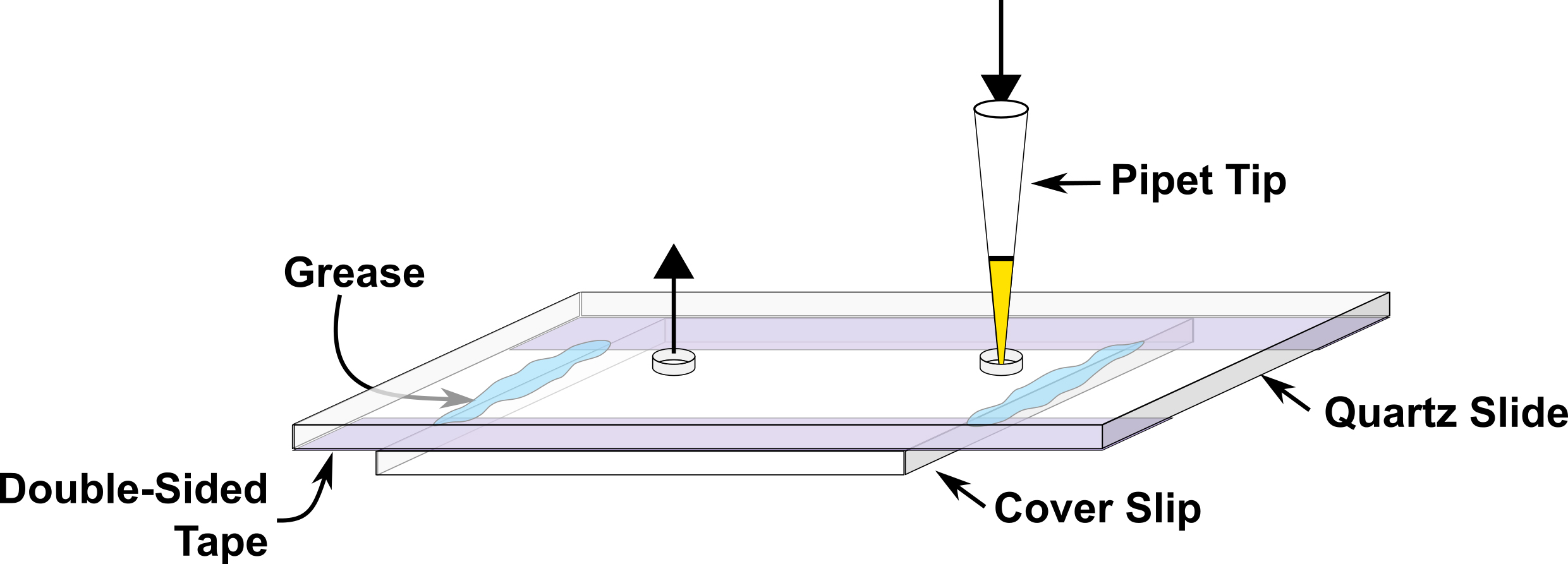
Figure 2. Illustration of the microfluidic sample chamber. Buffers or sample solutions are introduced through the entrance port via a pipette tip, as shown. Any excess solution flows out of the exit port, as shown. - Fill the sample chamber with 0.2 mg/ml neutravidin and incubate for five minutes followed by final buffer wash.
- Fill the sample chamber with 100 μl of receptor-nanodisc complexes (approximately 1 nM concentration), incubate for 10 min to allow complexes (containing 5% biotinylated lipids) to bind to neutravidin on the surface, then flush the chamber with imaging buffer.
- Clean quartz slides and cover slips, passivate the surfaces with polyethylene glycol (5% biotinylated), as described previously (Lamichhane et al., 2010).
- Single-molecule fluorescence data acquisition
- A microscope capable of total internal reflection fluorescence (TIRF) measurements, equipped with a 532 nm (green) laser for excitation (prism-based or objective-based illumination) and an intensified charge coupled device (CCD) camera capable of detecting the fluorescence emission from single fluorophores (Berezhna et al., 2012) is used to record a TIRF image of the surface at regular time intervals (e.g., every 100 msec or shorter, depending on the frame time of the CCD camera) for a total period of at least 100 sec. Many discrete fluorescent spots should be visible in the image, each representing individual receptor-nanodisc complexes (Figure 3A).
- Add a ligand, such as the agonist formoterol or inverse agonist ICI 118,551 (1 mM), to the sample chamber, allow ligand to interact with receptor (30 min to 1 h) and repeat the data acquisition. Alternatively, prepare a new sample chamber containing a pre-incubated mixture of receptor-nanodisc complexes and ligand.
- As a control to test for non-specific binding, add receptor-nanodisc complexes to a surface lacking neutravidin. Very few fluorescent spots should be visible on the surface (Figure 3B).
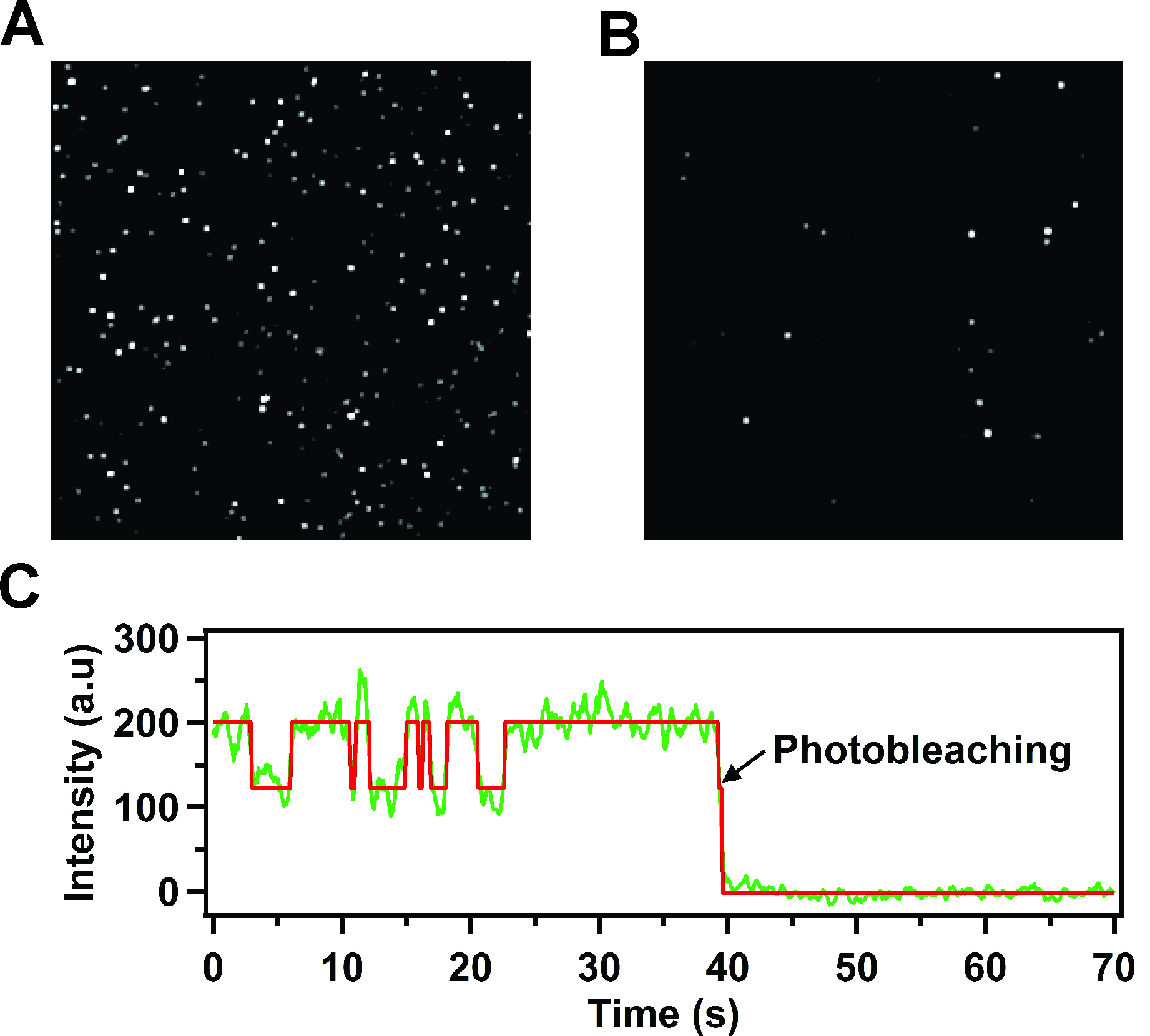
Figure 3. TIRF images of immobilized receptor-nanodisc complexes and representative time trace for a single receptor-nanodisc complex (reproduced from Lamichhane et al., 2015). A. Typical TIRF image showing a 5 x 5 μm region (approximate dimensions) of immobilized Cy3-labeled β2AR reconstituted in nanodiscs. Each spot is due to the fluorescence emitted from an individual receptor-nanodisc complex. The very bright spots are due to a small fraction of receptor-nanodisc aggregates, which are readily identified and excluded from the data analysis. B. Corresponding TIRF image for a control surface lacking neutravidin. The number of fluorescent spots is greatly reduced relative to part A, indicating a negligible level of non-specific adsorption of receptor-nanodisc complexes. C. Representative fluorescence intensity versus time trace from a single nanodisc-bound receptor, showing repeated two-state intensity jumps prior to an irreversible photobleaching event. The red line is the best fit from Hidden Markov modeling. The single-step photobleaching transition confirms that a single receptor molecule is contained within the nanodisc.
- A microscope capable of total internal reflection fluorescence (TIRF) measurements, equipped with a 532 nm (green) laser for excitation (prism-based or objective-based illumination) and an intensified charge coupled device (CCD) camera capable of detecting the fluorescence emission from single fluorophores (Berezhna et al., 2012) is used to record a TIRF image of the surface at regular time intervals (e.g., every 100 msec or shorter, depending on the frame time of the CCD camera) for a total period of at least 100 sec. Many discrete fluorescent spots should be visible in the image, each representing individual receptor-nanodisc complexes (Figure 3A).
Data analysis
- Use data acquisition software (available from https://cplc.illinois.edu/software/) to acquire the emission intensity of each identified spot on the surface at each time point.
- Filter the data to reject spots that: (a) show very high emission intensity, indicative of receptor-nanodisc aggregates, or (b) show photoblinking transitions (emission intensity transiently samples a zero-intensity state). For subsequent analysis, select time traces that show a single-step photobleaching transition, indicative of a single receptor molecule (Figure 3B).
- For each selected time trace, establish the background intensity level after the single photobleaching transition. Correct the intensity at all preceding time points accordingly. Steps 2 and 3 (Data analysis) can be automated using suitable MATLAB scripts.
- The majority of selected time traces should show reversible transitions between two distinct intensity levels (Figure 3C). Using at least 100 traces, construct binned intensity histograms representing the intensity states sampled in the entire collection (using Igor software). The histograms typically reveal two distinct peaks. Fit the histograms with two Gaussian functions to identify the centers and relative areas of each peak (using Igor or Origin software).
- Repeat this analysis for the data obtained in the presence of agonist or inverse agonist ligands. The intensity peak representing the active state of the receptor will be enhanced in the presence of agonist, while the peak representing the inactive state will be enhanced in the presence of inverse agonist, thereby establishing the identities of the two peaks. The peak areas report the equilibrium distribution of inactive and active conformational states of the receptor.
- Fit each selected intensity trace using Hidden Markov analysis (McKinney et al., 2006) to determine the dwell times spent in each state prior to transition to the other (example of fit shown in Figure 3C). Construct histograms of the dwell times for each state from the entire collection of complexes. Fit the histograms with single-exponential decay functions to determine rate constants for the activation or deactivation transitions (using Igor software). If the single-exponential fits are poor, based on the reduced chi-square value, refit the histograms with bi-exponential functions (Lamichhane et al., 2015).
Recipes
- Low salt wash buffer
10 mM HEPES, pH = 7.5, 20 °C
10 mM MgCl2
20 mM KCl
2 mM EDTA - High salt wash buffer
10 mM HEPES, pH = 7.5, 20 °C
10 mM MgCl2
20 mM KCl
1 M NaCl - Solubilization buffer
100 mM HEPES, pH = 7.5, 20 °C
300 mM NaCl
1% (w/v) DDM, 0.2% CHS (w/v) - Wash buffer 1
50 mM HEPES, pH = 7.5, 20 °C
800 mM NaCl
10 mM MgCl2
20 mM imidazole
0.05% (w/v) DDM, 0.01% (w/v) CHS
8 mM ATP - Wash buffer 2
50 mM HEPES, pH = 7.5, 20 °C
150 mM NaCl
20 mM imidazole
0.05% (w/v) DDM, 0.01% (w/v) CHS - Labeling buffer
50 mM HEPES, pH = 7.5, 20 °C
150 mM NaCl
0.05% (w/v) DDM, 0.01% (w/v) CHS
GPCR ligand - SEC elution buffer
50 mM PBS, pH = 7.4, 20 °C - IMAC elution buffer
50 mM HEPES, pH = 7.5, 20 °C
150 mM NaCl
200 mM imidazole
0.05% (w/v) DDM, 0.01% (w/v) CHS - Final buffer
50 mM HEPES, pH = 7.5, 20 °C
150 mM NaCl - Imaging buffer (final buffer plus oxygen scavenging system)
50 mM HEPES, pH = 7.5, 20 °C
150 mM NaCl
2 mM trolox
2-5% (w/v) glucose
50 µg/ml glucose oxidase
10 µg/ml catalase
Acknowledgments
This work was supported by the Roadmap Initiative grant P50 GM073197 from the National Institutes of Health. The protocol described here is adapted from the work of Lamichhane et al. (2015).
References
- Baker, J. G. (2010). The selectivity of beta-adrenoceptor agonists at human β1-, β2- and β3-adrenoceptors. Br J Pharmacol 160(5): 1048-1061.
- Bayburt, T. H. and Sligar, S. G. (2010). Membrane protein assembly into Nanodiscs. FEBS Lett 584(9): 1721-1727.
- Berezhna, S. Y., Gill, J. P., Lamichhane, R. and Millar, D. P. (2012). Single-molecule Forster resonance energy transfer reveals an innate fidelity checkpoint in DNA polymerase I. J Am Chem Soc 134(27): 11261-11268.
- Cherezov, V., Liu, J., Griffith, M., Hanson, M. A. and Stevens, R. C. (2008). LCP-FRAP assay for pre-screening membrane proteins for in meso crystallization. Cryst Growth Des 8(12): 4307-4315.
- Cherezov, V., Rosenbaum, D. M., Hanson, M. A., Rasmussen, S. G., Thian, F. S., Kobilka, T. S., Choi, H. J., Kuhn, P., Weis, W. I., Kobilka, B. K. and Stevens, R. C. (2007). High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318(5854): 1258-1265.
- Hwang, H., Kim, H. and Myong, S. (2011). Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc Natl Acad Sci U S A 108(18): 7414-7418.
- Lamichhane, R., Liu, J. J., Pljevaljcic, G., White, K. L., van der Schans, E., Katritch, V., Stevens, R. C., Wuthrich, K. and Millar, D. P. (2015). Single-molecule view of basal activity and activation mechanisms of the G protein-coupled receptor β2AR. Proc Natl Acad Sci U S A 112(46): 14254-14259.
- Lamichhane, R., Solem, A., Black, W. and Rueda, D. (2010). Single-molecule FRET of protein-nucleic acid and protein-protein complexes: surface passivation and immobilization. Methods 52(2): 192-200.
- Liu, J. J., Horst, R., Katritch, V., Stevens, R. C. and Wuthrich, K. (2012). Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science 335(6072): 1106-1110.
- McKinney, S. A., Joo, C. and Ha, T. (2006). Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J 91(5): 1941-1951.
- Rasmussen, S. G., Choi, H. J., Fung, J. J., Pardon, E., Casarosa, P., Chae, P. S., Devree, B. T., Rosenbaum, D. M., Thian, F. S., Kobilka, T. S., Schnapp, A., Konetzki, I., Sunahara, R. K., Gellman, S. H., Pautsch, A., Steyaert, J., Weis, W. I. and Kobilka, B. K. (2011). Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469(7329): 175-180.
- Ritchie, T. K., Grinkova, Y. V., Bayburt, T. H., Denisov, I. G., Zolnerciks, J. K., Atkins, W. M. and Sligar, S. G. (2009). Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol 464: 211-231.
- Roth, C. B., Hanson, M. A. and Stevens, R. C. (2008). Stabilization of the human β2-adrenergic receptor TM4-TM3-TM5 helix interface by mutagenesis of Glu122(3.41), a critical residue in GPCR structure. J Mol Biol 376(5): 1305-1319.
- Stennett, E. M., Ciuba, M. A., Lin, S. and Levitus, M. (2015). Demystifying PIFE: The photophysics behind the protein-induced fluorescence enhancement phenomenon in Cy3. J Phys Chem Lett 6(10): 1819-1823.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lamichhane, R., Liu, J. J., Pauszek III, R. F. and Millar, D. P. (2017). Fluorophore Labeling, Nanodisc Reconstitution and Single-molecule Observation of a G Protein-coupled Receptor. Bio-protocol 7(12): e2332. DOI: 10.21769/BioProtoc.2332.
Category
Biochemistry > Protein > Single-molecule Activity > Imaging
Biochemistry > Protein > Labeling
Biochemistry > Protein > Structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









