- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Functional Analysis of Connexin Channels in Cultured Cells by Neurobiotin Injection and Visualization
Published: Vol 7, Iss 11, Jun 5, 2017 DOI: 10.21769/BioProtoc.2325 Views: 8870
Reviewed by: Andrea GramaticaGunjan MehtaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantifying Intracellular Distributions of HaloTag-Labeled Proteins With SDS-PAGE and Epifluorescence Microscopy
Julia Shangguan and Ronald S. Rock
Jul 20, 2025 2510 Views

Fluorescence Lifetime-Based Separation of FAST-Labeled Cellular Compartment
Aidar R. Gilvanov [...] Yulia A. Bogdanova
Oct 5, 2025 1326 Views

Optimization of Adipogenic Differentiation Protocol for Murine and Human Cell Culture Models
Junwan Fan [...] Wenyan He
Jan 20, 2026 233 Views
Abstract
Functional gap junction channels between neighboring cells can be assessed by microinjection of low molecular weight tracer substances into cultured cells. The extent of direct intercellular communication can be precisely quantified by this method. This protocol describes the iontophoretic injection and visualisation of Neurobiotin into cultured cells.
Keywords: Gap junctionBackground
Gap junctions are intercellular conduits formed between neighboring cells, allowing the diffusional exchange of low molecular mass molecules (< 1.8 kDa). A gap junction channel consists of two hemichannels (connexons) docked to each other. Each connexon is a hexameric assembly of protein subunits termed connexins (Cx). The gap junction protein gene family consists of 20 members in mice and 21 in humans. Connexins are named according to their approximate molecular mass in kDa e.g., Cx43 has an approximate molecular mass of 43 kDa (for review see Söhl and Willecke, 2004). Different connexins are widely expressed in a variety of tissues throughout development where they mediate electrical as well as metabolic coupling. Furthermore, second messenger molecules and ions can be exchanged by direct diffusion through gap junctional channels.
Neurobiotin (N-(2-aminoethyl)biotinamide) is a compound of 286 Da molecular mass and a charge of +1 under physiological conditions. Due to its small size, this tracer passes even those gap junction channels which are not permeable to other common tracers of higher molecular mass e.g., Lucifer Yellow or carboxyfluorescein (Hampson et al., 1992), therefore representing a very sensitive method to detect gap junctional intercellular communication. Compared to the similar tracer biocytin, Neurobiotin appears to be superior regarding solubility, and stability. Furthermore, the compound can be selectively iontophoresed with positive current and subsequently fixed using paraformaldehyde or glutaraldehyde (Kita and Armstrong, 1991). As Neurobiotin does not show autofluorescence it needs to be detected using Avidin conjugated either to horseradish peroxidase or directly linked to a fluorescent dye.
Materials and Reagents
- 6 cm tissue culture treated culture dishes (several distributers available e.g., Corning, Tewksbury, MA)
- GB 100-F8P borosilicate glass capillaries (Science Products GmbH, Hofheim, Germany)
- Syringe (1 ml)
- Spinal needle (0.5 mm, 25 G)
- Reaction tube
- Cell line or primary cell preparation of interest
- pcDNATM3.1/Zeo(+)
- HistoGreen HRP-substrate Kit (Linaris, catalog number: E109 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9333 )
- Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S3264 )
- Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P9791 )
- Neurobiotin (Vector Laboratories, catalog number: SP-1120 )
- Rhodamine B isothiocyanate-Dextran (Sigma-Aldrich, catalog number: R9379 )
- Tris base (Sigma-Aldrich, catalog number: T1503 )
- Lithium chloride (LiCl) (Sigma-Aldrich, catalog number: L9650 )
- 50% glutaraldehyde solution (Sigma-Aldrich, catalog number: 340855 )
- Triton X-100 (Sigma-Aldrich, catalog number: X100 )
- Avidin D coupled horseradish peroxidase (Vector Laboratories, catalog number: A-2004 )
- PBS (see Recipes)
- Neurobiotin/Rhodamine B isothiocyanate-Dextran solution (see Recipes)
- LiCl solution (see Recipes)
- 0.5% glutaraldehyde solution (see Recipes)
- Triton X-100 solution (see Recipes)
- Avidin D coupled horseradish peroxidase solution (see Recipes)
Equipment
- Microelectrode-holder suitable for 1 mm glass capillaries with AgCl electrode (e.g., World Precision Instruments, model: MEH8 )
- Dual Microiontophoresis current generator SYS-260 (World Precision Instruments, FL)
- Zeiss IM35 inverted fluorescence microscope (Zeiss, model: Zeiss IM35 ) equipped with:
- Heated stage set to 37 °C
- HBO lamp (100 W)
- Appropriate filter set to detect Rhodamine B fluorescence (Ex/Em 570/590; Filter Set 20 HE)
- The microscope should be placed on an anti-vibration microscope desk
- Incubator
- Micropipette puller P97 (Sutter Instrument, model: P-97 )
- Micromanipulator Injectman (Eppendorf, Hamburg, Germany)
- AgCl reference electrode (disc)
Procedure
- Setup
Connect the micropipette holder to the positive and the reference electrode to the negative output of the microiontophoresis current generator. The microelectrode holder is mounted to the micromanipulator (Figure 1).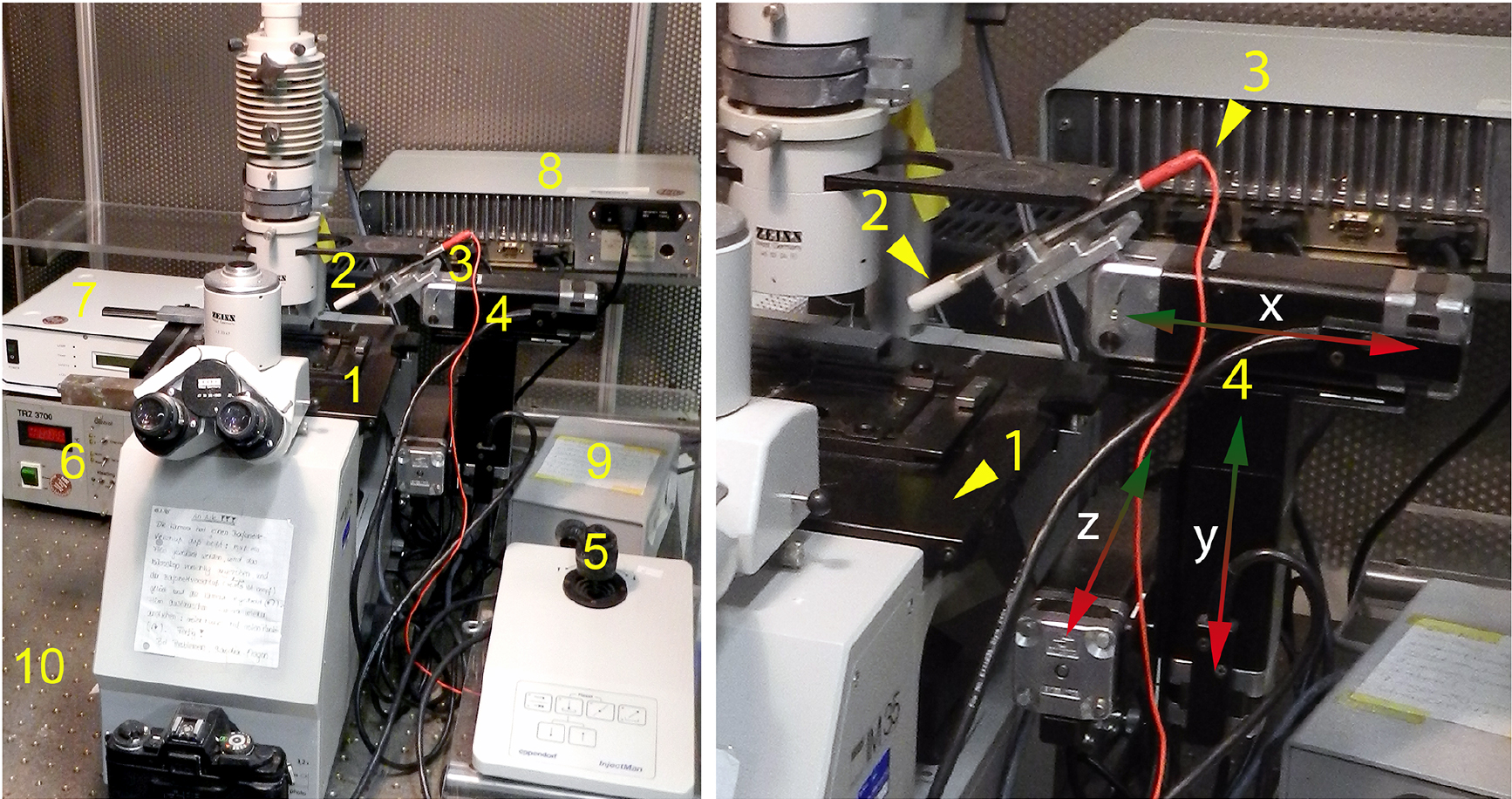
Figure 1. Experimental setup. We use a Zeiss IM35 inverted fluorescence microscope equipped with heating stage (set to 37 °C), HBO fluorescence lamp (100 W) and appropriate filter set to detect Rhodamine B fluorescence. The microscope is placed on an anti-vibration table. The microelectrode holder is mounted to the micromanipulator and connected to the positive output of the microiontophoresis current generator. The reference electrode is connected to the negative output of the current generator (reference electrode and current generator are not shown on this photograph). 1: heating stage, 2: rod with connection for micropipette holder, 3: positive wire (iontophoresis current generator -> micropipette) 4: micromanipulator (moving in x-, y- and z-axis), 5: controller (micromanipulator), 6: temperature controller (heating stage), 7: power supply (fluorescence lamp), 8: power supply (micromanipulator), 9: power supply (microscope lamp), 10: anti-vibration microscope desk. - Neurobiotin tracer microinjection
- Culture the cells on 6 cm cell culture dishes until they reach the desired confluency e.g., for HeLa cells we use 70-80% confluency.
Note: Neurobiotin microinjection can be performed with any adherent cell line or primary cell culture to determine the extent of gap junctional intercellular communication. - One hour prior to injection: Remove cell culture medium and wash the cells with PBS in order to remove cell debris. Aspirate the PBS and add 2 ml of fresh cell culture medium. Put the cells back into the incubator.
- Use the micropipette puller to manufacture injection capillaries with a tip size of 1-2 µm from GB 100-F8P borosilicate glass capillaries.
- Load the capillaries with Neurobiotin/Rhodamine B isothiocyanate-Dextran solution by bringing the backside of the micropipette in contact with the solution. The capillarity of the pipette will draw the fluid to the tip (Figure 2A).
- Fill the capillary with LiCl solution with a syringe by inserting a spinal needle into the backside of the capillary while withdrawing the needle as the micropipette fills. Avoid any air bubbles (Figure 2B). Little air bubbles can be removed by holding the micropipette tip down and gently flicking the pipette with the finger.
- Fill the micropipette holder with LiCl solution using syringe and needle. Avoid any air bubbles (Figure 2C).
- Carefully place the pipette into the micropipette holder. Avoid any air bubbles (Figure 2D).
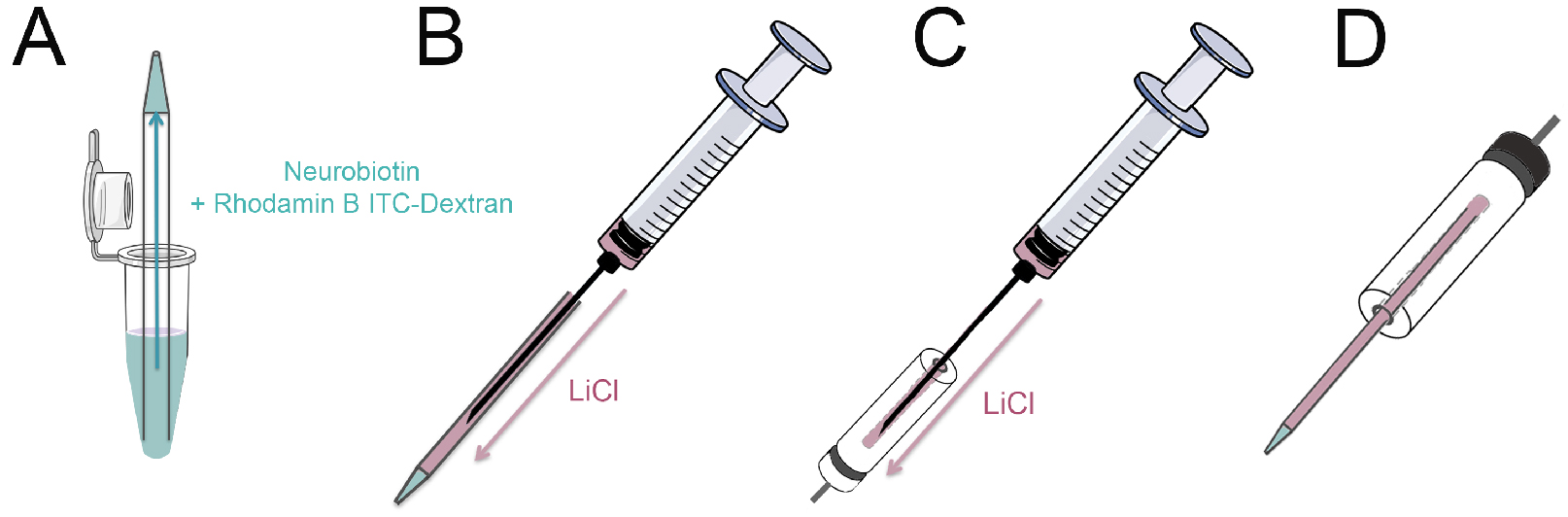
Figure 2. Loading the micropipette. A. Place the micropipette tip up into a 500 µl reaction tube containing 20 µl Neurobiotin/Rhodamine B isothiocyanate-Dextran solution. The capillarity of the pipette will draw the fluid to the tip. B. Fill the capillary with LiCl solution with a syringe by inserting a spinal needle into the backside of the capillary; C. Fill the micropipette holder with LiCl solution; D. Place the pipette into the micropipette holder. Avoid any air bubbles during all steps. Parts of this schematic were adapted from Servier Medical Art. - Put the 6 cm culture dish containing the cells under the microscope (heated stage set to 37 °C) and place the reference electrode into the culture medium. Set up a negative retaining current to prevent tracer leakage from the micropipette.
Note: Before starting microinjection, use a marker to draw a line at the bottom of the culture dish and place injections along this line. This facilitates going back to the injected cell under the microscope after staining. - While looking at the cells through the microscope at 10x magnification, use the micromanipulator to carefully position the tip of the micropipette next to the cell you want to inject. Penetrate the cell with the pipette. Be careful not to break the pipette tip at the bottom of the culture dish. This procedure needs some practice.
- Switch from brightfield to fluorescence microscopy. The pipette tip should get visible due to red Rhodamin B fluorescence (E: 590 nm).
Note: We recommend to work in a darkened room and keep main lights switched off during the experiment. This facilitates switching between brightfield and fluorescence microscopy. - Inject the tracer solution by applying a positive current of 20 nA for 10-15 sec. The injected cell should fill with the red fluorescent dye. The dye should not spread into neighboring cells as Rhodamine B isothiocyanate-Dextran is too big to pass gap junction channels (Figure 3A, Inset).
- Carefully retract the pipette tip from the cell.
- Repeat the procedure to inject 10-15 cells per 6 cm dish within max. 15 min.
- Culture the cells on 6 cm cell culture dishes until they reach the desired confluency e.g., for HeLa cells we use 70-80% confluency.
- Neurobiotin tracer visualisation
- Wash the injected cells twice with PBS directly after the last injection.
- Fix the cells with glutaraldehyde solution for 5 min at room temperature.
- Wash three times with PBS.
- Permeabilize cells overnight at 4 °C using Triton X-100 solution.
- Wash cells three times with PBS.
- Incubate cells for 90 min at room temperature with Avidin D coupled horseradish peroxidase solution.
- Wash three times with PBS.
- Use the HistoGreen HRP-substrate Kit to indirectly visualize Neurobiotin. The use of HistoGreen-substrate will label cells which received the Neurobiotin tracer in blue (green) color (Figures 3A-3C).
Notes:- Alternatively to the HistoGreen HRP-substrate kit, 3,3’-Diaminobenzidine (DAB) staining can be used for visualization.
- Monitor the staining reaction by looking at the cells under the microscope to avoid overstaining. If the staining reaction should get too intense even after short incubation times, dilute the Avidin D coupled horse radish peroxidase solution (Recipe 6) or reduce the amount of injected Neurobiotin by shortening the injection time (step B11).
- Alternatively to the HistoGreen HRP-substrate kit, 3,3’-Diaminobenzidine (DAB) staining can be used for visualization.
- The number of coupled cells can be determined by manual counting of labeled cells.
- Wash the injected cells twice with PBS directly after the last injection.
Data analysis
For data analysis, we take photographs of every successfully injected cell using phase contrast microscopy (Figures 3A-3C). Some injections may fail due to breakage of the microcapillary tip or due to overstaining and are excluded from the analysis. We exclude an injection from analysis if the staining is so intense that single cells can no longer be discriminated from each other. The extent of gap junctional intercellular communication (GJIC) can be assessed by manual counting of labeled cells. We suggest to inject at minimum three individual 6 cm plates (try to perform 10-15 injections per plate) and count at least 8-10 injections on each plate to get a representative result. Usually, we set the average number of coupled cells in the control sample (e.g., cells that have not been treated with pharmacological gap junction inhibitors) to 100% coupling and compare the other results to this (Figure 3D). As an example, representative data from two different experiments are presented:
- HeLa cells transfected with an expression plasmid (pcDNATM3.1/Zeo(+)) carrying the Cx43 ORF were injected with Neurobiotin. The tracer (blue color) was visualised using the HistoGreen HRP-substrate Kit. The white asterisk marks the injected cell. A spread of the tracer into neighboring cells can be observed (Figure 3A). The inset shows the fluorescence of Rhodamine B isothiocyanate-Dextran which was co-injected with Neurobiotin. The tip of the micropipette as well as the injected cell are visible (Figure 3A inset). The HeLa cell line used for these experiment does not show endogenous connexin expression. Therefore, a spread of the tracer is not observed when mock transfected HeLa cells are injected with Neurobiotin (Figure 3B).
- HM-1 mouse embryonic stem cells (HM-1 mESCs, Magin et al., 1992) were injected with Neurobiotin. mESCs endogenously express the gap junction proteins Cx31, Cx43 and Cx45 (Wörsdörfer et al., 2008 and 2016) (Figure 3C). Pre-incubation of mESCs with the pharmacological gap junction inhibitor 18-alpha glycyrrethinic acid concentration dependently (0-100 µm) decreased the number of coupled cells as assessed by Neurobiotin microinjection (Figure 3D).
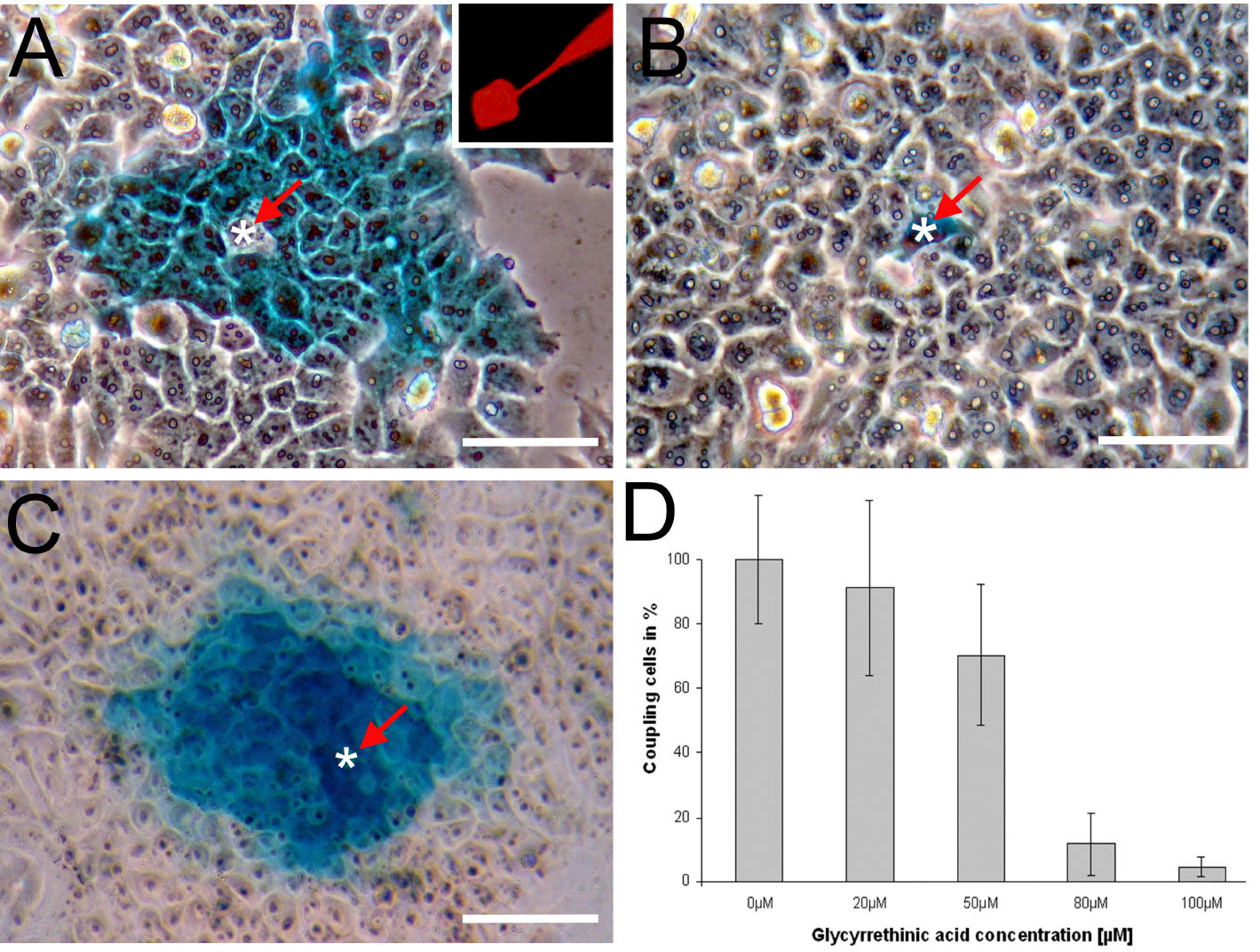
Figure 3. Microinjection of Neurobiotin. A. HeLa cells transfected with Cx43 were injected with Neurobiotin. The white asterisk marks the injected cell. The inset shows the fluorescence of Rhodamine B isothiocyanate-Dextran. B. The parental HeLa cell line used for this experiment does not show gap junctional intercellular communication; C. Mouse embryonic stem cells (mESCs) injected with Neurobiotin; D. Pre-incubation of mESCs with the gap junction inhibitor 18-alpha glycyrrethinic acid concentration dependently (0-100 µm) decreased the number of coupled cells. The number of coupling mESCs in the absence of the gap junction inhibitor was defined as 100%. Scale bars = 50 µm.
Notes
- In order to confirm the specificity of the assay the following control experiments should be performed:
- Monitor Neurobiotin microinjection by looking at the Rhodamine B fluorescence. The co-injected Rhodamine B isothiocyanate–Dextran is too big to pass gap junction channels and should not spread into neighboring cells.
- Cells can be incubated with pharmacological gap junction inhibitors (e.g., 18-alpha glycyrrethinic acid or its derivate carbenoxolone, Sigma-Aldrich, St. Louis, MO) prior to Neurobiotin microinjection. This should concentration dependently decrease the spread of dye into neighboring cells (Davidson et al., 1988; Figure 3D). Be aware that not all connexins are sensitive to these gap junction blockers: for example, Cx43 is sensitive to 18-alpha glycyrrethinic, while Cx31 and Cx45 are not (He et al., 2005; Wörsdörfer et al., 2008).
Recipes
- PBS
133 mM NaCl
2.7 mM KCl
8.1 mM Na2HPO4
1.5 mM KH2PO4
in (dd)H2O
pH 7.2 - Neurobiotin/Rhodamine B isothiocyanate-Dextran solution (20 µl per experiment)
6% (w/v) Neurobiotin
0.4% (w/v) Rhodamine B isothiocyanate-Dextran
0.1 M Tris base
in (dd)H2O
pH 7.6
Store at 4 °C, protect from light - LiCl solution
Note: Prepare 5-10 ml per experiment.
1 M LiCl in (dd)H2O - 0.5% glutaraldehyde solution
1% (v/v) 50% glutaraldehyde solution in PBS (final concentration 0.5%) - Triton X-100 solution
2% Triton X-100 in PBS - Avidin D coupled horseradish peroxidase solution
0.1% in PBS
Prepare freshly before the experiment
Acknowledgments
Work in Bonn laboratory was supported by the German Research Foundation SFB645 project B2 and the Bonn Forum in Biomedicine to K.W. We thank Katharina Günther for proofreading the manuscript. Parts of the schematic procedure shown in Figure 2 were produced using the image bank of Servier Medical Art (www.servier.com) licensed under a Creative Commons Attribution 3.0 Unported License. The injection of Neurobiotin as a tracer for gap junctional communication between cultured cells has been often used in the literature. An excellent, extensive review on the permeability of connexin channels has been published by Harris and Locke (2009).
References
- Davidson, J. S. and Baumgarten, I. M. (1988). Glycyrrhetinic acid derivatives: a novel class of inhibitors of gap-junctional intercellular communication. Structure-activity relationships. J Pharmacol Exp Ther 246(3): 1104-1107.
- Hampson, E. C., Vaney, D. I. and Weiler, R. (1992). Dopaminergic modulation of gap junction permeability between amacrine cells in mammalian retina. J Neurosci 12(12): 4911-4922.
- Harris, A. L. and Locke, D. (2009). Permeability of connexin channels. In: Harris and Locke (Eds.). Connexins: A guide. Humana Press.
- He, L. Q., Cai, F., Liu, Y., Liu, M. J., Tan, Z. P., Pan, Q., Fang, F. Y., Liang, D. S., Wu, L. Q., Long, Z. G., Dai, H. P., Xia, K., Xia, J. H. and Zhang, Z. H. (2005). Cx31 is assembled and trafficked to cell surface by ER-Golgi pathway and degraded by proteasomal or lysosomal pathways. Cell Res 15(6): 455-464.
- Kita, H. and Armstrong, W. (1991). A biotin-containing compound N-(2-aminoethyl)biotinamide for intracellular labeling and neuronal tracing studies: comparison with biocytin. J Neurosci Methods 37(2): 141-150.
- Magin, T. M., McWhir, J. and Melton, D. W. (1992). A new mouse embryonic stem cell line with good germ line contribution and gene targeting frequency. Nucleic Acids Res 20(14): 3795-3796.
- Söhl, G. and Willecke, K. (2004). Gap junctions and the connexin protein family. Cardiovasc Res 62(2): 228-232.
- Wörsdörfer, P., Bosen, F., Gebhardt, M., Russ, N., Zimmermann, K., Komla Kessie, D., Sekaran, T., Egert, A., Ergün, S., Schorle, H., Pfeifer, A., Edenhofer, F. and Willecke, K. (2016). Abrogation of gap junctional communication in ES cells results in a disruption of primitive endoderm formation in embryoid bodies. Stem Cells 35(4): 859-871.
- Wörsdörfer, P., Maxeiner, S., Markopoulos, C., Kirfel, G., Wulf, V., Auth, T., Urschel, S., von Maltzahn, J. and Willecke, K. (2008). Connexin expression and functional analysis of gap junctional communication in mouse embryonic stem cells. Stem Cells 26(2): 431-439.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wörsdörfer, P. and Willecke, K. (2017). Functional Analysis of Connexin Channels in Cultured Cells by Neurobiotin Injection and Visualization. Bio-protocol 7(11): e2325. DOI: 10.21769/BioProtoc.2325.
Category
Stem Cell > Adult stem cell > Maintenance and differentiation
Cell Biology > Cell imaging > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










