- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Generation of Mutant Pigs by Direct Pronuclear Microinjection of CRISPR/Cas9 Plasmid Vectors
Published: Vol 7, Iss 11, Jun 5, 2017 DOI: 10.21769/BioProtoc.2321 Views: 13094
Reviewed by: Longping Victor TseFang XuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Primary Neuronal Culture and Transient Transfection
Shun-Cheng Tseng [...] Eric Hwang
Jan 20, 2025 2780 Views

Reprogramming White Fat Cells for Adipose Manipulation Transplantation (AMT) Therapy
Kelly An [...] Nadav Ahituv
Aug 5, 2025 2271 Views

Generation of Insulin-Producing Alpha TC1-6 Cells Using EpiCRISPR System for Targeted DNA Methylation
Marija B. Đorđević [...] Melita S. Vidaković
Oct 20, 2025 1278 Views
Abstract
A set of Cas9 and single guide CRISPR RNA expression vectors was constructed. Only a very simple procedure was needed to prepare specific single-guide RNA expression vectors with high target accuracy. Since the de novo zygotic transcription had been detected in mouse embryo at the 1-cell stage, the plasmid DNA vectors encoding Cas9 and GGTA1 gene specific single-guide RNAs were micro-injected into zygotic pronuclei to confirm such phenomenon in 1-cell pig embryo. Our results demonstrated that mutations caused by these CRISPR/Cas9 plasmids occurred before and at the 2-cell stage of pig embryos, indicating that besides the cytoplasmic microinjection of in vitro transcribed RNA, the pronuclear microinjection of CRISPR/Cas9 DNA vectors provided an efficient solution to generate gene-knockout pig.
Keywords: CRISPR/Cas9Background
Since the initial discovery of a highly conserved 29 base pairs (bp) sequence tandemly repeated with a spacing of 32 bp downstream of the iap gene in Escherichia coli genome (Ishino et al., 1987; Nakata et al., 1989), a family of short regularly spaced repeats, varying in size from 25 to 50 bp, were found in about 50% of bacteria and 90% of archaea (Makarova et al., 2015). According to their characteristic structures, the name clustered regularly interspaced short palindromic repeats (CRISPRs) were introduced by Mojica (Mojica et al., 2009) and Jansen (Jansen et al., 2002) and are currently in general use. A set of CRISPR associated genes, cas1 to cas4, was first identified flanking the CRISPR loci by nucleotide sequence alignments (Jansen et al., 2002). New members of cas genes were identified in different bacterial species. According to the cas members within each host, the CRISPR-Cas systems can be classified into three major types based on the hallmark genes (Makarova et al., 2011; Burmistrz and Pyrc, 2015). The function of the CRISPR-Cas system was demonstrated by a seminar experiment. After being challenged by virulent bacteriophages: phage 858 and phage 2,972, new repeat-spacer units were observed on the leading end of the CRISPR array in the surviving Streptococcus thermophlis host cells. The DNA sequences of newly acquired spacers were matched to corresponding fragments, named proto-spacers, in the phage genomes. Streptococcus thermophlis strains with phage spacer(s) were resistant to the phage infection, while strains without phage spacer(s) were sensitive to the phage infection (Barrangou et al., 2007). To distinguish between the spacer in the host bacterial genome and the proto-spacer, which has the same sequence as the spacer in the invader genome, a proto-spacer adjacent motif (PAM) was evolved (Mojica et al., 2009; Shah et al., 2013). The CRISPR-Cas immunity was revealed, it can be divided into three stages (Rath et al., 2015; Wright et al., 2016). The first adaptation or acquisition stage is responsible for the acquisition of spacers into CRISPR array following the exposure to foreign mobile genetic elements, such as phages or plasmids. A Cas2 homodimer was sandwiched by two Cas1 homodimers forming a heterohexameric complex for all three types of CRISPR-Cas systems (Sternberg et al., 2016). In the second stage, the promoter embedded within the AT-rich leader sequence upstream of the CRISPR array transcribed the precursor CRISPR RNAs (pre-crRNAs), these were further processed into short CRISPR RNA (crRNA) guides by Cas proteins. Cas6 was involved in the RNA processing step in both of the type I and type III CRISPR-Cas systems (Charpentier et al., 2015; Hochstrasser and Doudna, 2015). Accompanied by the Cas9 protein, a trans-activating crRNA (tracrRNA) which contains an anti-repeat segment for duplex formation with the repeat compartment of crRNA was involved in the maturation of the crRNAs in the type II system (Deltcheva et al., 2011). In the last interference stage, in cooperation with a mature crRNA and a cascade of Cas proteins, the signature proteins Cas3 and Cas10 were integrated into the RNA guided endonuclease complex in the type I and type III CRISPR-Cas systems, respectively. The type II effector is simply composed of a Cas9 protein, a pair of processed mature crRNA and tracrRNA (Gasiunas et al., 2012). The crRNA and tracrRNA in the ternary complex can be substituted by a fused crRNA-tracrRNA single-guide RNA (sgRNA) (Jinek et al., 2012). Because of extreme simplicity, the Cas9-sgRNA, now commonly termed as CRISPR/Cas9, binary complexes were immediately applied in the field of gene editing (Cong et al., 2013; Mali et al., 2013).
Swine share a number of anatomic and physiologic characteristics with humans. Systems that are mostly cited as suitable models include cardiovascular, urinary, integumentary, and digestive system (Swindle et al., 2012). Therefore, pigs are considered as a good source of organs for xenotransplantation. Two strategies are currently utilized to overcome the interspecies rejection hurdles. The first one is to block donor organs expressing the antigens causing hyper-acute rejection, such as galactose-α1,3-galactose, N-glycolylneuroaminic acid and β1,4-N-acetylgalactosamine, by targeting the GGTA1, CMAH and β4GalNT2 genes, respectively (Estrada et al., 2015; Cooper et al., 2016). The second strategy is to prepare organs which are composed of acceptor’s cells in a surrogate animal by a blastocyst complementation technique (Kobayashi et al., 2010; Usui et al., 2012; Matsunari et al., 2013). To produce gene-knockout pigs, the traditional method uses gene editing in somatic cells, such as fetal fibroblasts, and somatic cell nuclear transfer (SCNT) techniques to create knockout zygotes. The examples of combining CRISPR/Cas9 and SCNT are reported to generate IgM JH (Chen et al., 2015), RUNX3 (Kang et al., 2016b), IL2RG (Kang et al., 2016a), and GGTA1/CMAH/β4GalNT2 triple knockout pigs (Estrada et al., 2015). Direct microinjection of DNA or RNA is another choice to prepare gene knockout pigs. Since the burst of de novo transcription in porcine zygotes was reported at the 4-cell stage (Anderson et al., 1999), RNA is preferred for micro-injection into embryos at the 1-cell stage. Previous studies have demonstrated that cytoplasmic microinjection of Cas9 mRNA with sgRNAs can produce Mitf (Wang et al., 2015) and DJ-1/Parkin/PINK1 triple-gene knockout pigs (Wang et al., 2016). Because de novo zygotic transcription had only been reported in the 1-cell stage mouse embryo (Ram and Schultz, 1993; Bouniol et al., 1995; Aoki et al., 1997), a trial was reported to produce GGTA1 knockout pigs by cytoplasmic microinjection of a CRISPR/Cas9 plasmid. With this strategy, CRISPR/Cas9 was expressed at, or later, than the 2-cell stage, and mosaic mutations on GGTA1 gene were found (Petersen et al., 2016). Pronucleus microinjection is needed to interpret whether zygotic transcription occurs at the 1-cell stage of pig embryo (Chuang et al., 2016).
A series of Cas9 and sgRNA expression vectors was constructed as shown in Figure 1. pCX-Flag2-NLS1-Cas9-NLS2, pCX-HA-NLS1-Cas9-NLS2, and pCX-Myc-NLS1-Cas9-NLS2 can be used to express Cas9 in mammalian cells. Three common tags are available to monitor Cas9 expressions. (Figure 1A) Porcine U6 promoter which could effectively derive short hairpin RNA [Chuang et al., 2009] was used to construct the ppU6-(BsaI)2-gRNA vector (Figure 1B) (Su et al., 2015). A pair of primers containing spacer and part of CRISPR repeat sequences, as shown in Figure 1B, is needed for each target site. Because guanine (G) is favored for U6 promoter as the first transcribed nucleotide, only proto-spacers initiated with G were chosen in our recent works.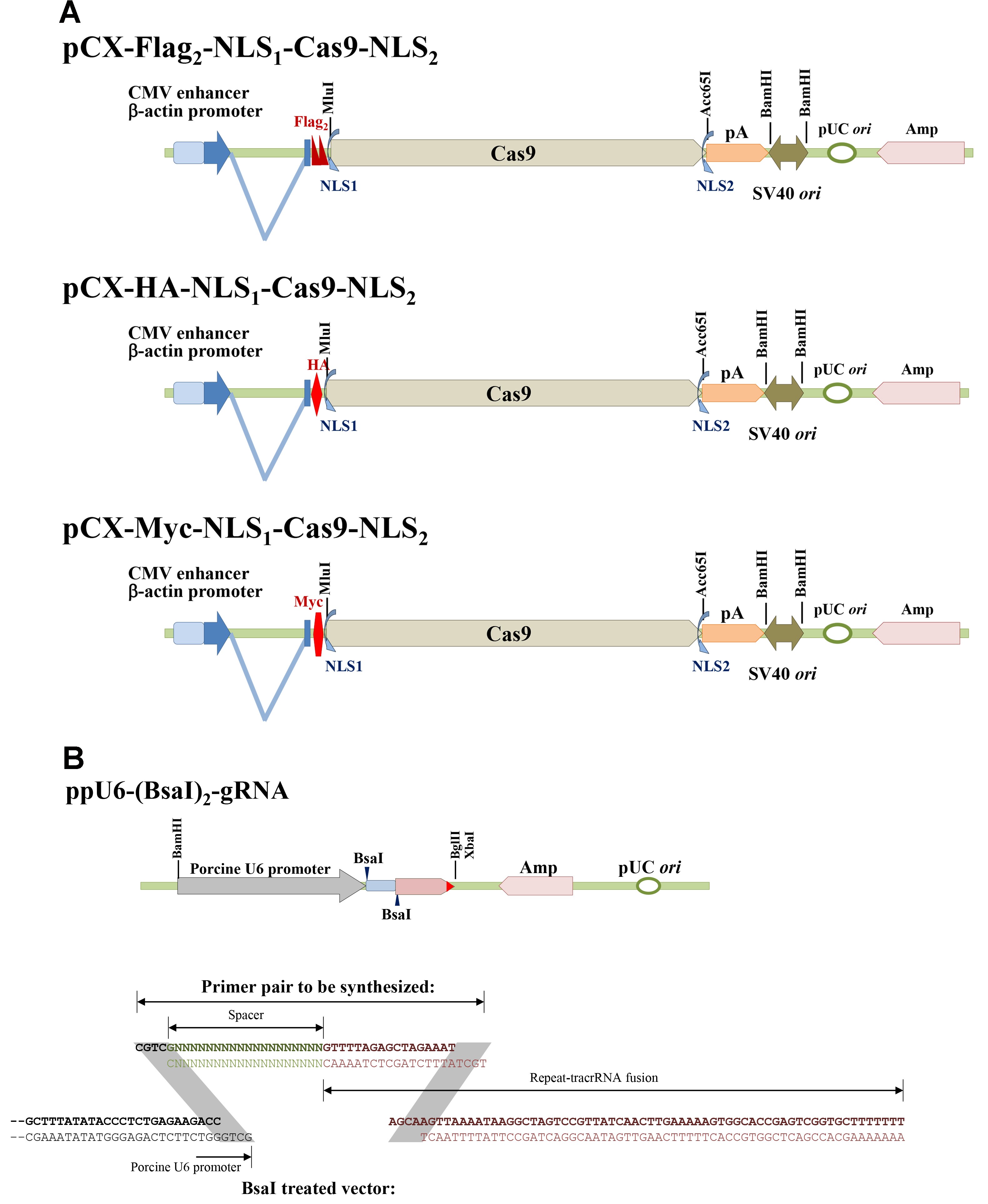
Figure 1. Scheme of the Cas9 and single-guide RNA expression vectors. A. pCX-Flag2-NLS1-Cas9-NLS2, pCX-HA-NLS1-Cas9-NLS2, and pCX-Myc-NLS1-Cas9-NLS2; B. ppU6-(BsaI)2-gRNA and primer pair designation.
Materials and Reagents
- Preparations of sgRNA expression vectors
- BsaI (New England Biolabs, catalog number: R0535 )
- T4 DNA ligase (Promega, catalog number: M1801 )
- Clean and Gel Extraction Kit (Biokit, catalog number: Bio-C300 )
- 2x ligation buffer (Promega, catalog number: C671A )
- pGEM-T Easy TA-cloning kit (Promega, catalog number: A1360 )
- Plasmid Miniprep Kit (Biokit, catalog number: Bio-P300 )
- Genomic DNA isolation Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: K0721 )
- NeonTM Transfection System 100 µl Kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: MPK10096 )
- Cesium chloride (CsCl) (Avantor® Performance Materials, J.T. Baker®, catalog number: 4042-02 )
- Tris-HCl, pH 8.0
- EDTA, pH 8.0
- Sodium chloride (NaCl)
- TE buffer (see Recipes)
- TEN buffer (see Recipes)
- BsaI (New England Biolabs, catalog number: R0535 )
- Collection of pig embryos
- Glass tube for embryo flashing (glass tube with outside diameter of 4 mm as shown in Figure 2)
- Falcon tube, 50 ml (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 339653 )
- Regumate® (containing 0.4% Altrenogest, Intervet, MSD, France, brand name: Regumate®)
- PMSG (pregnant mare serum gonadotropin) (ASFK Pharmaceutical, Japan, brand name: PMSG)
- hCG (human chorionic gonadotropin) (ASKA Pharmaceutical, Japan, brand name: hCG)
- PGF2α (prostaglandin F2α; Estrumate injection) (Intervet Deutschland, Germany)
- D-PBS (GE Healthcare, HyCloneTM, catalog number: SH30028.02 )
- Fetal bovine serum (FBS) (GE Healthcare, HyCloneTM, catalog number: SH30071.03 )
- Glass tube for embryo flashing (glass tube with outside diameter of 4 mm as shown in Figure 2)
- Embryo transfer
- 3M paper tape (3M, catalog number: 200-24mm )
- Atropine sulfate (TAI YU CHEMICAL & PHARMACEUTICAL)
- Stresnil (azaperona, 40 mg/ml) (Janssen Pharmaceutica N.V., Belgium, brand name: Stresnil)
- Citosol (Thiamylal sodium) (Shinlin Sinseng Pharmaceutical, Taiwan, brand name: Citosol)
- Amoxicillin (ampicillin, 150 mg/ml) (China Chemical & Pharmaceutical, CCPG, catalog number: E000853 )
- Heparin sodium (China Chemical & Pharmaceutical, CCPG, Taiwan, brand name: AGGLUTEX INJECTION)
- 3M paper tape (3M, catalog number: 200-24mm )
Equipment
- Preparations of sgRNA erxpression vectors
- Ultracentrifugator (Beckman Coulter, model: Optima XL-80 ) equipped with NVTi65 rotor
- Microcentrifugator (KUBOTA, model: 3740 )
- Dry bath incubator (Major Science, model: MD-01N )
- Thermocycler (Thermo Fisher Scientific, Applied BiosystemsTM, model: 2720 )
- Ultracentrifugator (Beckman Coulter, model: Optima XL-80 ) equipped with NVTi65 rotor
- Semen assessment
- computer-assisted sperm analysis system (UltiMate CASA system, Hamilton-Thorne Research, Beverly, MA)
- computer-assisted sperm analysis system (UltiMate CASA system, Hamilton-Thorne Research, Beverly, MA)
- Microinjection
- Centrifugator (KUBOTA, model: 8920 )
Note: This product has been discontinued. - Microcentrifugator (KUBOTA, model: 3740 )
- Inverted Differential Interference Contrast microscope (Olympus, model: IX71 )
- Capillary injection needle (Sutter Instrument, catalog number: BF100-78-10 )
- Capillary injection needle puller (Sutter Instrument, model: P-97 )
- Micromanipulator (NARISHGE, model: ON3-99D )
- Injectors (NARISHGE, model: IM-9B )
- Centrifugator (KUBOTA, model: 8920 )
- Collection of pig embryos and embryo transfer
- Operation table (NEWPORT, LW3048B-OPT, VIBRATION ISOLATION TABLE)
- Surgery mechanic devises (NARISHIGE, JAPAN)
- Operation table (NEWPORT, LW3048B-OPT, VIBRATION ISOLATION TABLE)
Procedure
- Preparations of sgRNA expression vectors
- Take 10 μg of ppU6-(BsaI)2-gRNA and let it be digested with 50 U of BsaI in 100 μl NEB3.1 buffer at 37 °C for 2 h.
- Purify the linearized vector DNA with a clean-up kit and elute it with 30 μl of 1/10x TE. Store the purified linearized vector at -20 °C before use.
- Dissolve the primers listed in Table 1 in TE and adjust the concentrations to 50 μM.
Notes:- Add each 10 μl of forward and reverse primers (Table 1, 50 μM, each) to a vial and mix them with 30 μl of TEN. Incubate the mixture at 90 °C for 10 min in a dry block heater.
- Transfer the mixture to a 65 °C dry block and incubate for 30 min. Then let the block gradually cool down to room temperature for about 30 min. Put the annealed primer pair on ice until the following ligation reaction.
Table 1. Primers for ppU6-sgRNA expression vector construction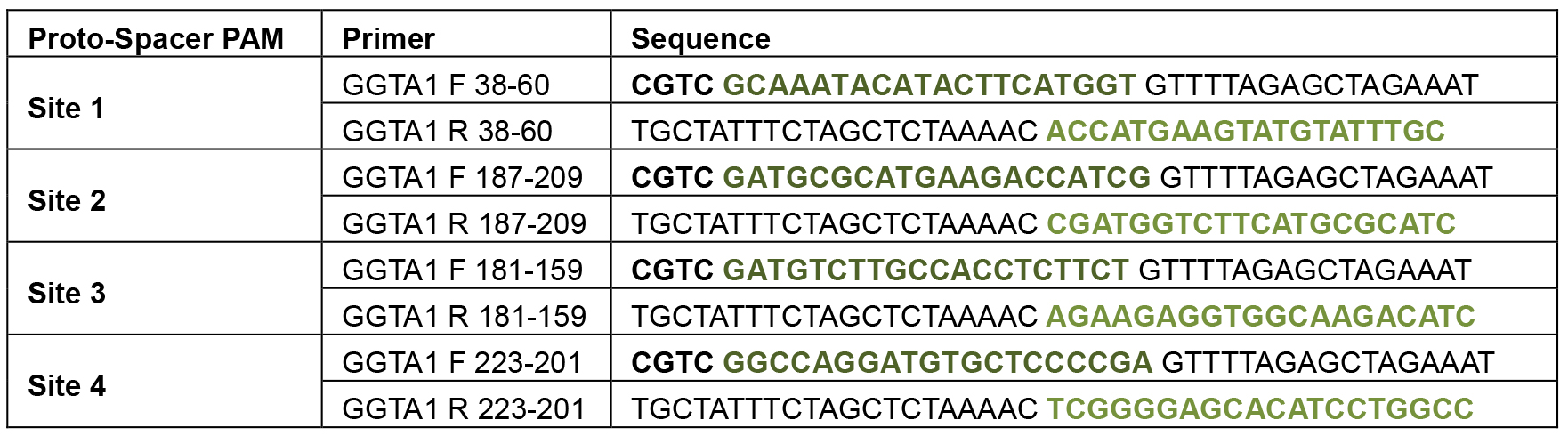
The first nucleotide of the last exon of pig GGTA1 gene was numbered as +1, and the last nucleotide of the last intron was numbered as -1 (Su et al., 2015).
- Add each 10 μl of forward and reverse primers (Table 1, 50 μM, each) to a vial and mix them with 30 μl of TEN. Incubate the mixture at 90 °C for 10 min in a dry block heater.
- For the ligation reaction, mix 1 μl of BsaI digested vector, 3 μl of annealed primer pair, 5 μl of 2x ligation buffer and 1 μl T4 DNA ligase in a vial on ice. Then incubate the vial at 16 °C for 90 min to perform the ligation reaction.
- After the transformation, isolate plasmid samples from the individual colonies. Use M13 reverse primer for DNA sequencing (Note 1).
- Purify the DNA for pronuclear microinjection by two rounds of CsCl density-gradient ultra-centrifugation. The NVTi65 rotor is used in general and the centrifugation conditions are set at 218,000 x g for 20 h at 20 °C.
- Take 10 μg of ppU6-(BsaI)2-gRNA and let it be digested with 50 U of BsaI in 100 μl NEB3.1 buffer at 37 °C for 2 h.
- Animals and animal care
- Raise all animals in a specific pathogen free farm. Feed the animals on a restricted diet (4% body weight) with free access to water.
- Raise the recipients normally but treat them with special care particularly during farrowing.
- All animals should be managed and treated in accordance to your federal and institutional guidelines.
- Raise all animals in a specific pathogen free farm. Feed the animals on a restricted diet (4% body weight) with free access to water.
- Synchronization and superovulation
- Synchronize the donors and recipients by feeding with a commercial ration supplemented with Regumate® (containing 0.4% Altrenogest; Intervet, MSD, France) for 15 days to synchronize their estrus cycles. Feed the recipients with Regumate® for one more day.
- On the last day of feeding Regumate®, inject PGF2 (250 μg Cloprostenol) intramuscularly (i.m.) to lyse any remaining corpus luteum.
- For superovulation induction, inject all pigs intramuscularly with PMSG and hCG to induce follicle growth/oocyte maturation and ovulation, respectively. Briefly, on the next day of the last feeding with Regumate®, inject all of the donors with 2,000 i.u. of PMSG. Then inject them with 1,500 i.u. of hCG 78 h later. Inject the recipients by the same regimen with 1,500 i.u. PMSG and 1,250 i.u. hCG to induce follicle growth/oocyte maturation and ovulation, respectively.
- For artificial insemination, artificially inseminate the donors with fresh diluted semen containing at least 3 billion viable sperms with 80% mobility as assessed by computer-assisted sperm analysis system at 24 h and 36 h after hCG injection. Induce the synchronized ovulation of recipients by the same methods with a 12 h delay and without insemination.
- Synchronize the donors and recipients by feeding with a commercial ration supplemented with Regumate® (containing 0.4% Altrenogest; Intervet, MSD, France) for 15 days to synchronize their estrus cycles. Feed the recipients with Regumate® for one more day.
- Pronuclear oocytes recovery
- During 54-56 h after hCG injection, sacrifice all of the donors to harvest the upper uterus horns and oviducts.
- For the collection of zygotes, insert and connect infundibulum with a glass tube to guide the D-PBS into a 50 ml Falcon tube. Flush out the fertilized eggs from the oviducts by injection of 20 ml D-PBS with 20% fetal calf serum (FCS) into the fallopian tube near the uterotubal junction. (Figure 2)
- Keep the embryos in DPBS in a thermal box at 37 °C and carry them to a laboratory for further micromanipulation within 30 min.
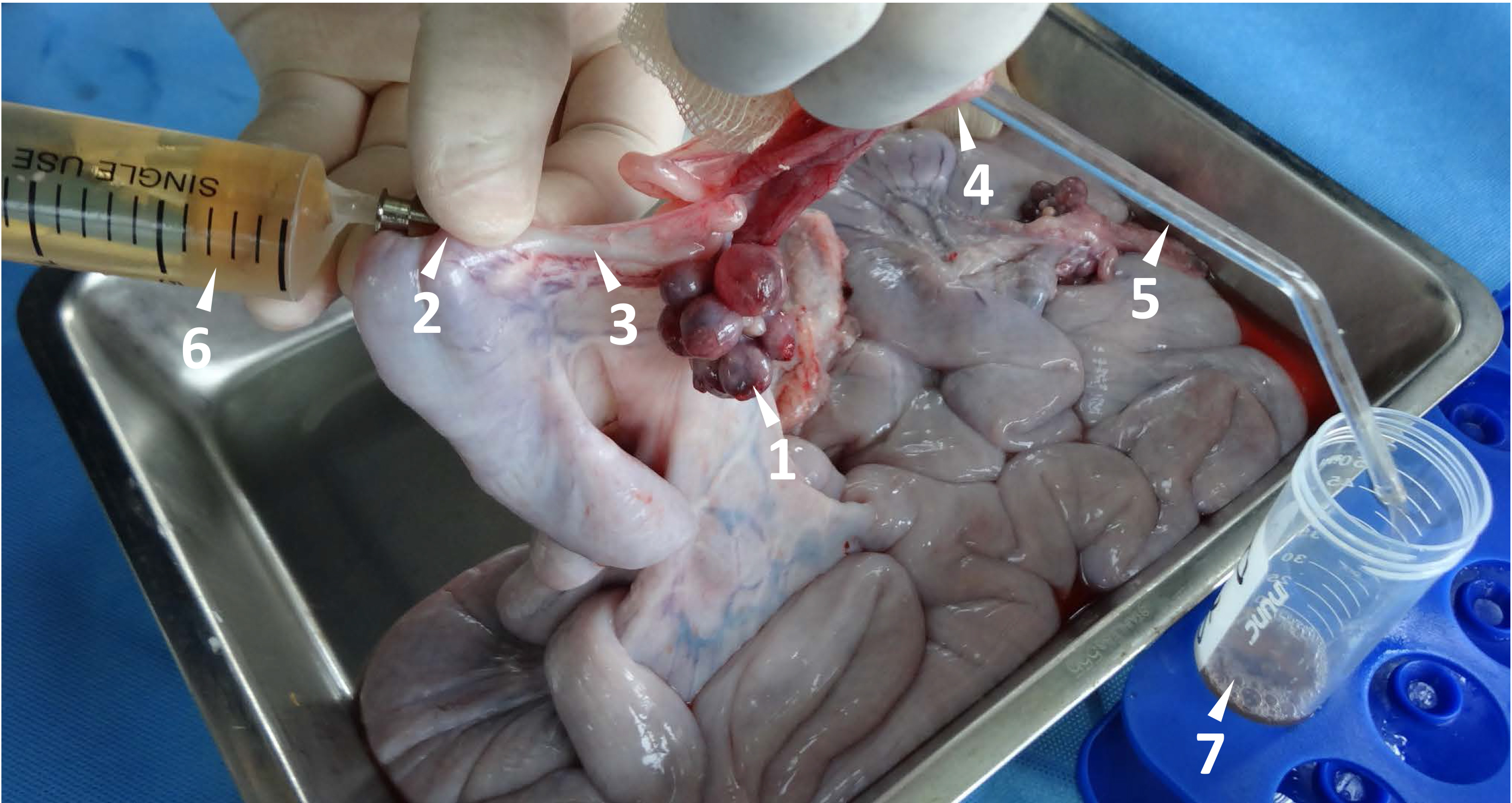
Figure 2. Flushing newly fertilized eggs from porcine fallopian tube. The maturation and ovulation are indicated by the follicle with an ovulation point (arrow head 1). A stainless needle (#19) with a blunt end is used to penetrate the uterus wall, and it is inserted through the uterotubal junction (arrow head 2) into the fallopian tube (arrow head 3). A glass tube (arrow hear 5, outside diameter 4 mm) is connected into the other end of the fallopian tube through the open of infundibulum (arrow head 4). The fertilized eggs within the fallopian tube are flushed out by 20 ml D-PBS with a syringe (arrow head 6) and collected in a 50 ml Falcon tube (arrow head 7).
- During 54-56 h after hCG injection, sacrifice all of the donors to harvest the upper uterus horns and oviducts.
- Microinjection
- Centrifuge the recovered fertilized eggs at 15,000 x g for 10 to 15 min at 25 °C to expose their pronuclei (Figure 3).
Notes:- Then put the embryos into a 20 µl D-PBS micro-drop on an inverted microscope equipped the differential interference contrast system.
- Perform the pronuclear microinjection under 300-fold magnification.
- Hold the embryos in a proper position to reveal a clear view of pronuclei.
- Then put the embryos into a 20 µl D-PBS micro-drop on an inverted microscope equipped the differential interference contrast system.
- Then microinject the CRISPR/Cas9 DNA mixture into one pronucleus through a capillary needle. The diameter of tip opening for an injection pipette is about 0.5 µm. After applying injection pressure, the continuous flow stream of DNA is shown by moving the zygote away from the tip of the microinjection capillary. Before injection into pronuclei, the pipette tip was placed in the perivitelline space (PVS). A suitable injection flow speed is adjusted by increasing or decreasing the injection pressure according to the swelling velocity of PVS. The concentrations of plasmid DNA were 6 ng/µl for pCX-Flag2-NLS1-Cas9-NLS2 and 2 ng/µl for each of the four ppU6-sgRNA vectors (Note 2).
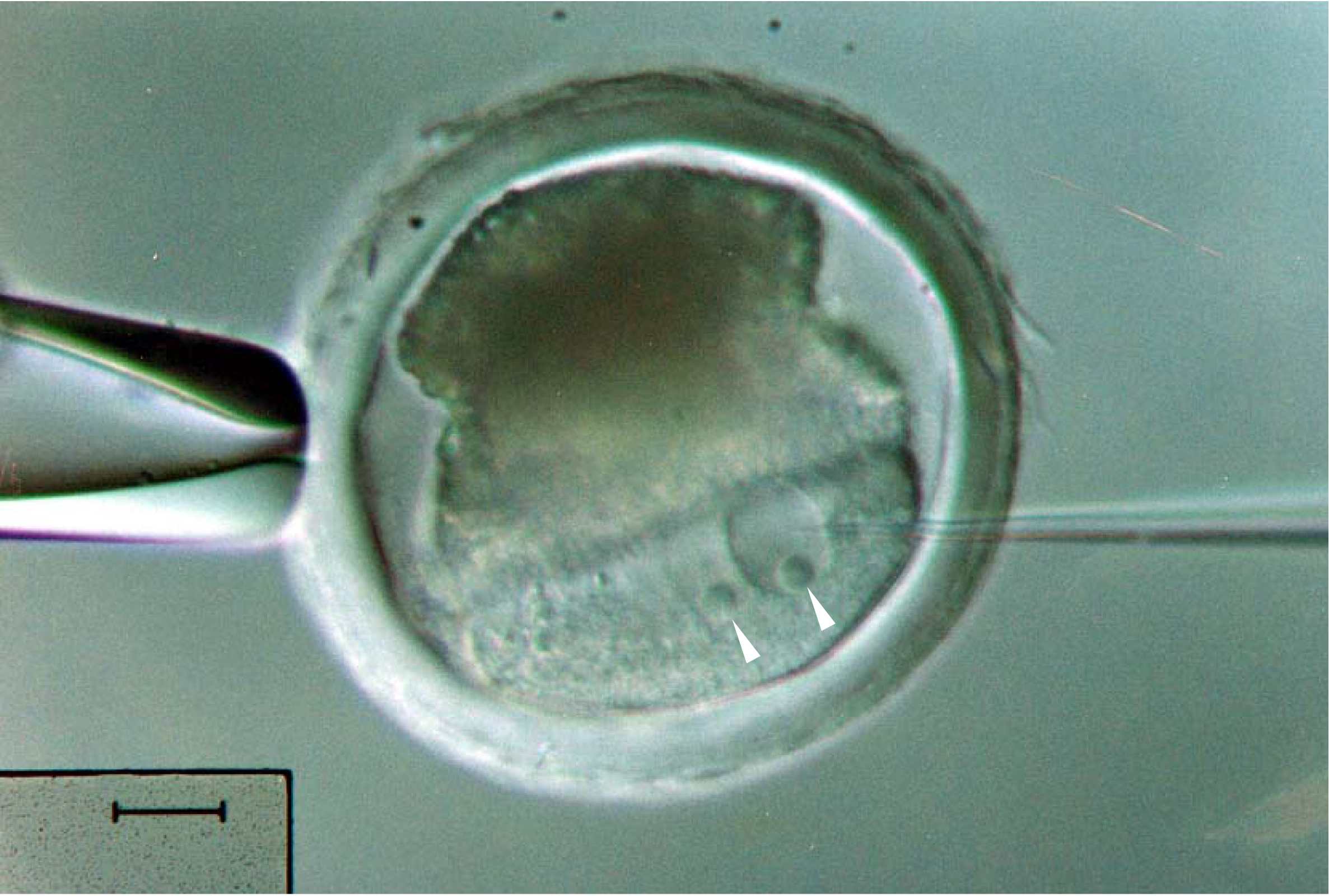
Figure 3. Pronuclear microinjection of a pig embryo. Centrifugation is critical to reveal the pronuclei, indicated by arrowheads, from the optically dense contents in the pig embryo. Scale bar = 20 μm.
- Centrifuge the recovered fertilized eggs at 15,000 x g for 10 to 15 min at 25 °C to expose their pronuclei (Figure 3).
- Embryo transfer
- After microinjection of CRISPR/Cas9 plasmid vectors into the pronuclei, transfer 20 to 30 embryos into the oviduct through a transfer pipette by the surgical method:
- Before operation, perform a 24 h fasting with free access to water for all of the recipients.
- Twenty minutes before surgery, inject 5 mg atropine sulfate and 400 mg Stresnil to decease saliva discharge and sedate the animals, respectively.
- Set up an ear vein catheter and fix it by 3M paper tape for i.v. injection of Citosol to maintain anesthesia, with the duration of operations lasting for 40 to 50 min.
- Load and fasten the recipients upside down on the operation table. Clean their abdomen skin and shave and sterilize the hair. Then cover the animal with a sterilized drape.
- Create an 8-10 cm window on the linea alba among the last 2nd to 3rd nipple, pull a single side of uterus horns, fallopian tubes and ovary outside the abdomen, and then count the number of corpus luteum on the ovary. Put back the largest part of the uterus horn immediately into cavum abdominis. Keep the remaining upper uterus horn, fallopian tube, infundibulum and ovary outside for embryo transfer (Figure 4A).
- Load 10-15 embryos in D-PBS in a glass transferring tube controlled by mouth pipet and transfer the embryos into the oviduct isthmus (Figure 4B).
- Rinse the uterus horn by saline containing heparin sulfate (50,000 U/L), and then put it back into the cavum abdominis.
- Before suturing the cut window, infuse 10 ml of amoxicillin into the cavum abdominis.
- After embryos transference, close the peritoneum parietale and muscle of the abdominal wall by continuous suture, and suture the adipose layer and skin by interrupted vertical mattress.
- Finally, inject 10 ml of amoxicillin i.m. at the neck during the following three days to prevent infection.
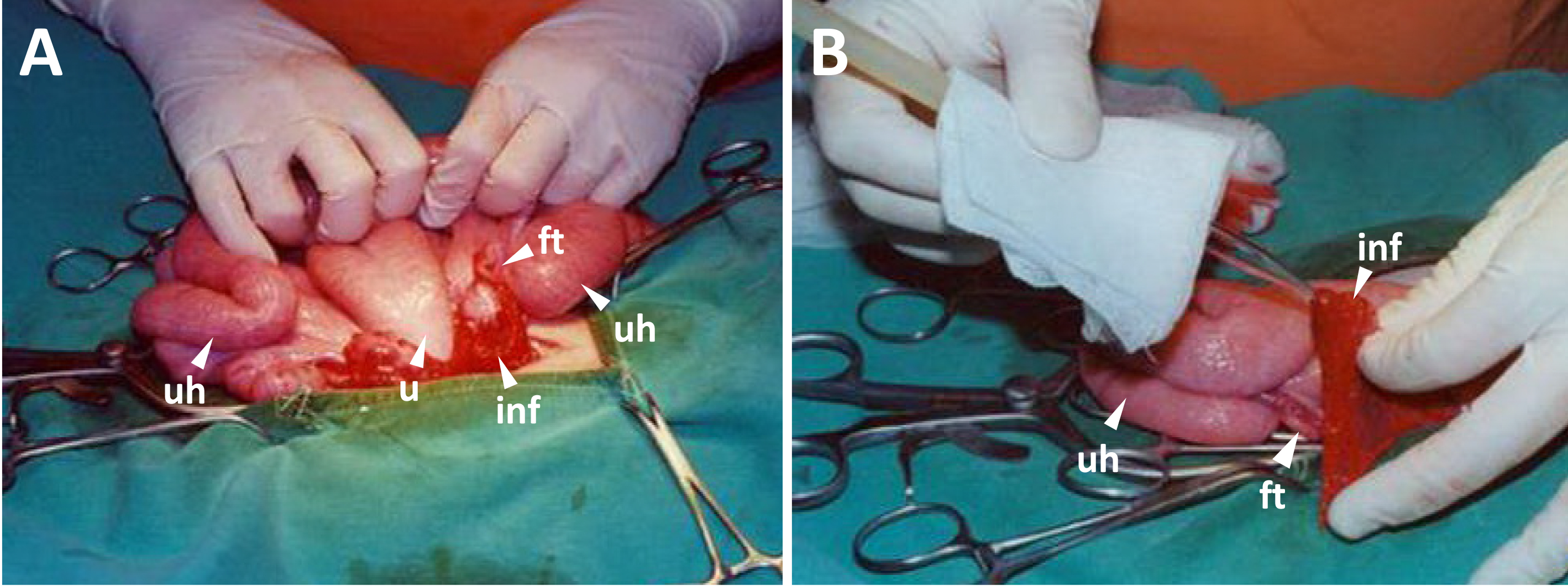
Figure 4. Embryo transfer. A. Upper uterus horns, uterus, fallopian tubes, infundibula and ovaries are kept outside for embryo transfer. B. The embryos are loaded in a glass tube controlled by mouth pipet, and transferred into the oviductal isthmus (abbreviations: u for the uterus, uh for the uterus horn, ft for the fallopian tube, and inf for the infundibulum).
- Farrowing and piglets care
- Move the sows to the farrowing pens seven days before the expected delivery day.
- Observe the nervous and restless behaviors of the sows; check the lactation further by hand milking.
- On the day when sows start to lactate, check the sows intensively to ascertain any one of them need assistance for delivering, and take care of the neonates.
- On the next day when farrowing was completed, collect the piglets’ tail tissue samples for DNA extraction and further genomic analysis.
- Move the sows to the farrowing pens seven days before the expected delivery day.
Data analysis
The genomic DNA samples isolated from pig tail tissues were analyzed by PCR with primers listed in Table 2. The PCR products were cloned into the pGEM-T Easy TA-cloning vector. Individual clones of plasmids were sequenced to interpret the mutations. In a typical experiment (Chuang et al., 2016), 41 pronucleus microinjected embryos were transferred into two surrogate sows. One of them was pregnant and 7 piglets were delivered. Two founders: L537-12 and L537-13, were detected carrying GGTA1 mutations in the tail tissues. Wild type DNA sequence and a 4 bp (TTGG) deletion at site 1 were revealed in the L537-12 sample. A 7 bp (TGGTTGG) deletion and another 7 bp (CATGGTT) deletion were identified in the L537-13 sample. After crossing with wild type mate, the 4 bp (TTGG) deletion was measured in 5 of the 14 offspring of the L537-12. The 7 bp (TGGTTGG) and 7 bp (CATGGTT) deletions were detected in 5 and 3, respectively, among the 13 offspring of L537-13. It is noteworthy that among the last 13 offspring, one piglet carried a 149 bp deletion (fragment between site 1 and site 4) which was not detected in the founder’s tail tissue. Theoretically, for a heterologous male, half of the sperms are mutants. For a 50% mosaic male, 25% of the sperms are mutants. As 5 of the 14 offspring of the L537-12 founder carry the 4 bp (GGTT) deletion, it indicates that the mosaic ratio is between 50 to 100%. Since 5 and 3 of the 13 offspring of the L537-13 founder carry the 7 bp (TGGTTGG) and 7 bp (CATGGTT) deletions, respectively, it also supports this conclusion. The other offspring of the L537-13 carrying the 149 bp deletion indicates that the deletion reaction occurred after the 4-cell embryonic stage. It is possible that the majority of the germline was just occupied by the descents of the mutated cells in the L537-12 and L537-13 cases. However, these data also supply a clue that GGTA1 mutations caused by CRISPR/Cas9 plasmids might occur before and at the 2-cell stage of pig zygotes.
Table 2. Primer pairs for PCR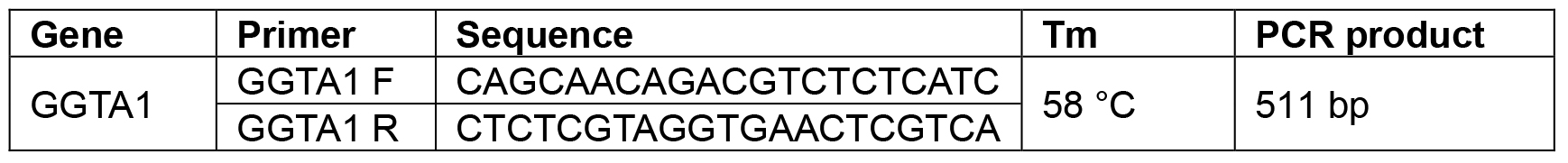
Notes
- If PAGE purified primers were used, more than 90% of clones analyzed were composed of correct sequences. E. coli JM109 competent cell was routinely used in our lab.
- The gene editing efficiencies of ppU6-GGTA138-60-gRNA, ppU6-GGTA1187-209-gRNA, ppU6-GGTA1181-159-gRNA, and ppU6-GGTA1223-201-gRNA, (site 1 to site 4, respectively) were first checked at cell level with LLC-PK1 cells (Su et al., 2015). Higher efficiencies were found by ppU6-GGTA1187-209-gRNA and ppU6-GGTA1223-201-gRNA. In this pronuclear microinjection experiment, all four ppU6-GGTA1-gRNA vectors were co-injected; coincidently, only indel mutations at site 1 and site 4 were detected.
Recipes
- TE buffer
10 mM Tris-HCl, pH 8.0
1 mM EDTA, pH 8.0 - TEN buffer
10 mM Tris-HCl, pH 8.0
1 mM EDTA, pH 8.0
0.5 M NaCl
Acknowledgments
This protocol is adapted from our recent works (Su et al., 2015; Chuang et al., 2016). We want to thank all the members who have been involved in these works. This work is supported by grants NSC 101-2313-B-059-001 to C.F.T. as well as MOST 104-2321-B-886-002 and MOST 105-2321-B-886-001 to C.K.C. from the Ministry of Science and Technology of Taiwan.
References
- Anderson, J. E., Matteri, R. L., Abeydeera, L. R., Day, B. N. and Prather, R. S. (1999). Cyclin B1 transcript quantitation over the maternal to zygotic transition in both in vivo- and in vitro-derived 4-cell porcine embryos. Biol Reprod 61(6): 1460-1467.
- Aoki, F., Worrad, D. M. and Schultz, R. M. (1997). Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 181(2): 296-307.
- Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., Romero, D. A. and Horvath, P. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315(5819): 1709-1712.
- Bouniol, C., Nguyen, E. and Debey, P. (1995). Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Exp Cell Res 218(1): 57-62.
- Burmistrz, M. and Pyrc, K. (2015). CRISPR-Cas systems in prokaryotes. Pol J Microbiol 64(3): 193-202.
- Charpentier, E., Richter, H., van der Oost, J. and White, M. F. (2015). Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol Rev 39(3): 428-441.
- Chen, F., Wang, Y., Yuan, Y., Zhang, W., Ren, Z., Jin, Y., Liu, X., Xiong, Q., Chen, Q., Zhang, M., Li, X., Zhao, L., Li, Z., Wu, Z., Zhang, Y., Hu, F., Huang, J., Li, R. and Dai, Y. (2015). Generation of B cell-deficient pigs by highly efficient CRISPR/Cas9-mediated gene targeting. J Genet Genomics 42(8): 437-444.
- Chuang, C. K., Chen, C. H., Huang, C. L., Su, Y. H., Peng, S. H., Lin, T. Y., Tai, H. C., Yang, T. S. and Tu, C. F. (2016). Generation of GGTA1 mutant pigs by direct pronuclear microinjection of CRISPR/Cas9 plasmid vectors. Anim Biotechnol: 1-8.
- Chuang, C. K., Lee, K. H., Fan, C. T. and Su, Y. S. (2009). Porcine type III RNA polymerase III promoters for short hairpin RNA expression. Anim Biotechnol 20(1): 34-39.
- Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P. D., Wu, X., Jiang, W., Marraffini, L. A. and Zhang, F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121): 819-823.
- Cooper, D. K., Ekser, B., Ramsoondar, J., Phelps, C. and Ayares, D. (2016). The role of genetically engineered pigs in xenotransplantation research. J Pathol 238(2): 288-299.
- Deltcheva, E., Chylinski, K., Sharma, C. M., Gonzales, K., Chao, Y., Pirzada, Z. A., Eckert, M. R., Vogel, J. and Charpentier, E. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471(7340): 602-607.
- Estrada, J. L., Martens, G., Li, P., Adams, A., Newell, K. A., Ford, M. L., Butler, J. R., Sidner, R., Tector, M. and Tector, J. (2015). Evaluation of human and non-human primate antibody binding to pig cells lacking GGTA1/CMAH/β4GalNT2 genes. Xenotransplantation 22(3): 194-202.
- Gasiunas, G., Barrangou, R., Horvath, P. and Siksnys, V. (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A 109(39): E2579-2586.
- Hochstrasser, M. L. and Doudna, J. A. (2015). Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci 40(1): 58-66.
- Ishino, Y., Shinagawa, H., Makino, K., Amemura, M. and Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169(12): 5429-5433.
- Jansen, R., Embden, J. D., Gaastra, W. and Schouls, L. M. (2002). Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43(6): 1565-1575.
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A. and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096): 816-821.
- Kang, J. T., Cho, B., Ryu, J., Ray, C., Lee, E. J., Yun, Y. J., Ahn, S., Lee, J., Ji, D. Y., Jue, N., Clark-Deener, S., Lee, K. and Park, K. W. (2016a). Biallelic modification of IL2RG leads to severe combined immunodeficiency in pigs. Reprod Biol Endocrinol 14(1): 74.
- Kang, J. T., Ryu, J., Cho, B., Lee, E. J., Yun, Y. J., Ahn, S., Lee, J., Ji, D. Y., Lee, K. and Park, K. W. (2016b). Generation of RUNX3 knockout pigs using CRISPR/Cas9-mediated gene targeting. Reprod Domest Anim 51(6): 970-978.
- Kobayashi, T., Yamaguchi, T., Hamanaka, S., Kato-Itoh, M., Yamazaki, Y., Ibata, M., Sato, H., Lee, Y. S., Usui, J., Knisely, A. S., Hirabayashi, M. and Nakauchi, H. (2010). Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell 142(5): 787-799.
- Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J., Charpentier, E., Horvath, P., Moineau, S., Mojica, F. J., Wolf, Y. I., Yakunin, A. F., van der Oost, J. and Koonin, E. V. (2011). Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9(6): 467-477.
- Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., Barrangou, R., Brouns, S. J., Charpentier, E., Haft, D. H., Horvath, P., Moineau, S., Mojica, F. J., Terns, R. M., Terns, M. P., White, M. F., Yakunin, A. F., Garrett, R. A., van der Oost, J., Backofen, R. and Koonin, E. V. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13(11): 722-736.
- Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., Norville, J. E. and Church, G. M. (2013). RNA-guided human genome engineering via Cas9. Science 339(6121): 823-826.
- Matsunari, H., Nagashima, H., Watanabe, M., Umeyama, K., Nakano, K., Nagaya, M., Kobayashi, T., Yamaguchi, T., Sumazaki, R., Herzenberg, L. A. and Nakauchi, H. (2013). Blastocyst complementation generates exogenic pancreas in vivo in apancreatic cloned pigs. Proc Natl Acad Sci U S A 110(12): 4557-4562.
- Mojica, F. J., Diez-Villasenor, C., Garcia-Martinez, J. and Almendros, C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155(3): 733-740.
- Nakata, A., Amemura, M. and Makino, K. (1989). Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. J Bacteriol 171(6): 3553-3556.
- Petersen, B., Frenzel, A., Lucas-Hahn, A., Herrmann, D., Hassel, P., Klein, S., Ziegler, M., Hadeler, K. G. and Niemann, H. (2016). Efficient production of biallelic GGTA1 knockout pigs by cytoplasmic microinjection of CRISPR/Cas9 into zygotes. Xenotransplantation 23(5): 338-346.
- Ram, P. T. and Schultz, R. M. (1993). Reporter gene expression in G2 of the 1-cell mouse embryo. Dev Biol 156(2): 552-556.
- Rath, D., Amlinger, L., Rath, A. and Lundgren, M. (2015). The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie 117: 119-128.
- Shah, S. A., Erdmann, S., Mojica, F. J. and Garrett, R. A. (2013). Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol 10(5): 891-899.
- Sternberg, S. H., Richter, H., Charpentier, E. and Qimron, U. (2016). Adaptation in CRISPR-Cas systems. Mol Cell 61(6): 797-808.
- Su, Y. H., Lin, T. Y., Huang, C. L., Tu, C. F. and Chuang, C. K. (2015). Construction of a CRISPR-Cas9 system for pig genome targeting. Anim Biotechnol 26(4): 279-288.
- Swindle, M. M., Makin, A., Herron, A. J., Clubb, F. J. Jr. and Frazier, K. S. (2012). Swine as models in biomedical research and toxicology testing. Vet Pathol 49(2): 344-356.
- Usui, J., Kobayashi, T., Yamaguchi, T., Knisely, A. S., Nishinakamura, R. and Nakauchi, H. (2012). Generation of kidney from pluripotent stem cells via blastocyst complementation. Am J Pathol 180(6): 2417-2426.
- Wang, X., Cao, C., Huang, J., Yao, J., Hai, T., Zheng, Q., Wang, X., Zhang, H., Qin, G., Cheng, J., Wang, Y., Yuan, Z., Zhou, Q., Wang, H. and Zhao, J. (2016). One-step generation of triple gene-targeted pigs using CRISPR/Cas9 system. Sci Rep 6: 20620.
- Wang, X., Zhou, J., Cao, C., Huang, J., Hai, T., Wang, Y., Zheng, Q., Zhang, H., Qin, G., Miao, X., Wang, H., Cao, S., Zhou, Q. and Zhao, J. (2015). Efficient CRISPR/Cas9-mediated biallelic gene disruption and site-specific knockin after rapid selection of highly active sgRNAs in pigs. Sci Rep 5: 13348.
- Wright, A. V., Nunez, J. K. and Doudna, J. A. (2016). Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164(1-2): 29-44.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chuang, C., Tu, C. and Chen, C. (2017). Generation of Mutant Pigs by Direct Pronuclear Microinjection of CRISPR/Cas9 Plasmid Vectors. Bio-protocol 7(11): e2321. DOI: 10.21769/BioProtoc.2321.
Category
Cell Biology > Cell engineering > CRISPR-cas9
Molecular Biology > DNA > Transfection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










