- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protein Localization in the Cyanobacterium Anabaena sp. PCC7120 Using Immunofluorescence Labeling
(*contributed equally to this work) Published: Vol 7, Iss 11, Jun 5, 2017 DOI: 10.21769/BioProtoc.2318 Views: 8322
Reviewed by: Dennis NürnbergElizabeth LibbyAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Rapid Detection of Proliferative Bacteria by Electrical Stimulation
Conor LA Edwards [...] Munehiro Asally
Feb 5, 2020 6565 Views

Tracking the Subcellular Localization of Surface Proteins in Staphylococcus aureus by Immunofluorescence Microscopy
Salvatore J. Scaffidi [...] Wenqi Yu
May 20, 2021 7812 Views

Calibrating Fluorescence Microscopy With 3D-Speckler (3D Fluorescence Speckle Analyzer)
Chieh-Chang Lin and Aussie Suzuki
Aug 20, 2024 1955 Views
Abstract
Techniques such as immunoflorescence are widely used to determine subcellular distribution of proteins. Here we report on a method to immunolocalize proteins in Anabaena sp. PCC7120 with fluorophore-conjugated antibodies by fluorescence microscopy. This method improves the permeabilization of cyanobacterial cells and minimizes the background fluorescence for non-specific attachments. In this protocol, rabbit antibodies were raised against the synthetic peptide of CyDiv protein (Mandakovic et al., 2016). The secondary antibody conjugated to the fluorophore Alexa488 was used due to its different emission range in comparison to the autofluorescence of the cyanobacterium.
Keywords: Cell divisionBackground
The immunofluorescence of cyanobacteria has been used extensively in cell identification and counting studies (Jin et al., 2016). However, immunolocalization of proteins has not been achieved efficiently in cyanobacteria. The most recurrent method to localize proteins is by fusing the protein of interest to a fluorescent protein such as GFP (Green Fluorescent Protein) that has a different emission wavelength (compared with cyanobacterial autofluorescence), and subsequent visualization using epifluorescence or confocal microscopy (Flores et al., 2016; Santamaria-Gomez et al., 2016).
The structural properties of cyanobacterial cells are the main challenges for applying immunofluorescence techniques. They consist of an inner membrane (IM), a peptidoglycan layer (PG) and an outer membrane (OM) (Rippka, 1988; Baulina, 2012; Jin et al., 2016), with an additional exopolysaccharide layer (sheath). The sheath is found in both unicellular and filamentous cyanobacteria (Kehr and Dittmann, 2015), and their thickness, composition and appearance depend on growth conditions, metabolic status, cell differentiation and other external and internal parameters (Jin et al., 2016). The sheath tends to trap antibodies by unspecific interactions. To avoid this problem, the washing and membrane permeabilization steps are the key to a successful immunofluorescence technique in cyanobacteria.
Materials and Reagents
- Pipette tips
- 1.5 ml tubes (Eppendorf)
- 50 ml tubes (Falcon tubes)
- Poly-L-lysine coated glass slides (Sigma-Aldrich, catalog number: P0425-72EA )
- Cover slips
- Petri dish
- Filter with a pore size of 0.2 µm
- Filamentous cyanobacterium, Anabaena sp. PCC7120
- BG-11 liquid supplied with 10 mM NaHCO3 (Rippka, 1988)
- Sodium hydrogen carbonate (NaHCO3) (EMD Millipore, catalog number: 106329 )
- Ethanol (EMD Millipore, catalog number: 1.00983.2500 )
- Triton X-100 (Winkler Limitada, catalog number: BM-2020 )
- Bovine serum albumin (BSA) (Divbio Science, catalog number: 41-903-100 )
- Tween-20 (Winkler Limitada, catalog number: TW-1652 )
- Secondary antibody Alexa Fluor 488 goat anti-rabbit IgG (Thermo Fisher Scientific, Invitrogen, catalog number: A11008 )
- ProLong Gold Antifade Mountant (Thermo Fisher Scientific, InvitrogenTM, catalog number: P36930 )
- Nail varnish
- Primary polyclonal antibody against All2320 peptide (Mandakovic et al., 2016)
- Sodium chloride (NaCl) (EMD Millipore, catalog number: 106404 )
- Potassium chloride (KCl) (EMD Millipore, catalog number: 104938 )
- Sodium dihydrogen phosphate (Na2HPO4) (EMD Millipore, catalog number: 106559 )
- Potassium phosphate monobasic (KH2PO4) (EMD Millipore, catalog number: 529568 )
- PBS buffer (pH 7.4) (see Recipes)
Equipment
- Pipettes
- Hydrophobic PAP pen (Thermo Fisher Scientific, catalog number: 008877 )
- Freezer at -20 °C
- Incubator at 4 °C
- Incubator at 55 °C
- Incubator at 24 °C with white light
- Olympus Fluoview FV1000 Confocal Microscope (Olympus, model: FluoviewTM FV1000 ) and objectives of 60x/1.35 NA oil immersion and 100x/1.40 NA oil immersion. Laserline Argon 488 (Excitation 495 nm, Emission 509 nm) and Laserline DPSS (Excitation 565 nm, Emission 590 nm)
- Moisture chamber (A dark plastic box with a moistened paper inside, PolarSafeTM Polypropylene Freezer Storage Box) (Argos Technologies, catalog number: R3130 )
Software
- ImageJ software (https://imagej.net)
Procedure
- Organism and growth conditions
- Anabaena sp. PCC7120 is grown axenically in BG-11 liquid medium at 24 °C under white light (25 µmol m-2 sec-1) and shaking at 90 rpm.
- Anabaena sp. PCC7120 is grown axenically in BG-11 liquid medium at 24 °C under white light (25 µmol m-2 sec-1) and shaking at 90 rpm.
- Fixation and permeabilization
- 50 µl of cyanobacterial culture (OD750 = 0.3) is added to a poly-lysine microscopy slide and dried for 20 min at 55 °C. Do not fix the cells with organic solvents or aldehydes.
- Fix the cell spots in 70% ethanol and incubate for 30 min at -20 °C. The slide is immersed in cold 70% ethanol contained in a Petri dish.
- The slides are air-dried for 20 min at room temperature.
- Use a hydrophobic PAP pen to draw a circle around the slide-mounted cell spot and let it dry for 15 min at room temperature.
- 50 µl of cyanobacterial culture (OD750 = 0.3) is added to a poly-lysine microscopy slide and dried for 20 min at 55 °C. Do not fix the cells with organic solvents or aldehydes.
- Labeling procedure
- Permeabilize the cells by adding a drop of 0.05% Triton X-100 in PBS for 2 min at room temperature, and repeat it three times by removing the drop each time with a pipette.
- Incubate with a drop of 3% BSA, 0.2% Triton X-100 in PBS for 1 h at 4 °C in a moisture chamber and remove this blocking solution.
- The cells are incubated with the primary antibody diluted 1:100 in a solution with 1% BSA, 0.05% Tween-20 in PBS. Pre-immune serum diluted 1:100 in a solution with 1% BSA, 0.05% Tween-20 in PBS was used as a control to ensure that the primary antibody is working. The cells with the solutions are incubated for 2 h at 4 °C, in a moisture chamber.
- Wash with 0.05% Triton X-100 in PBS for 2 min at room temperature, and repeat three times.
- Incubate with secondary antibody Alexa Fluor 488 goat anti-rabbit IgG (diluted in PBS with 1% BSA and 0.05% Tween-20, final concentration 10 µg/ml) for 45 min at 4 °C, in a moisture chamber.
- Wash with 0.05% Triton X-100 in PBS for 2 min at room temperature for three times.
- Add a drop of Prolong Antifade reagent to the sample slide, and then cover this with a cover slip while taking care not to create air bubbles. Seal with nail varnish.
- The slides are visualized with a Fluoview FV1000 Confocal Microscope and images are acquired in 16 bits. Alexa Fluor 488 is excited at a wavelength of 495 nm and emission is measured at 509 nm. To visualize autofluorescence due to phycobilisomes, samples are excited using 565 nm and fluorescence emission is monitored at 590 nm (Figure 1).
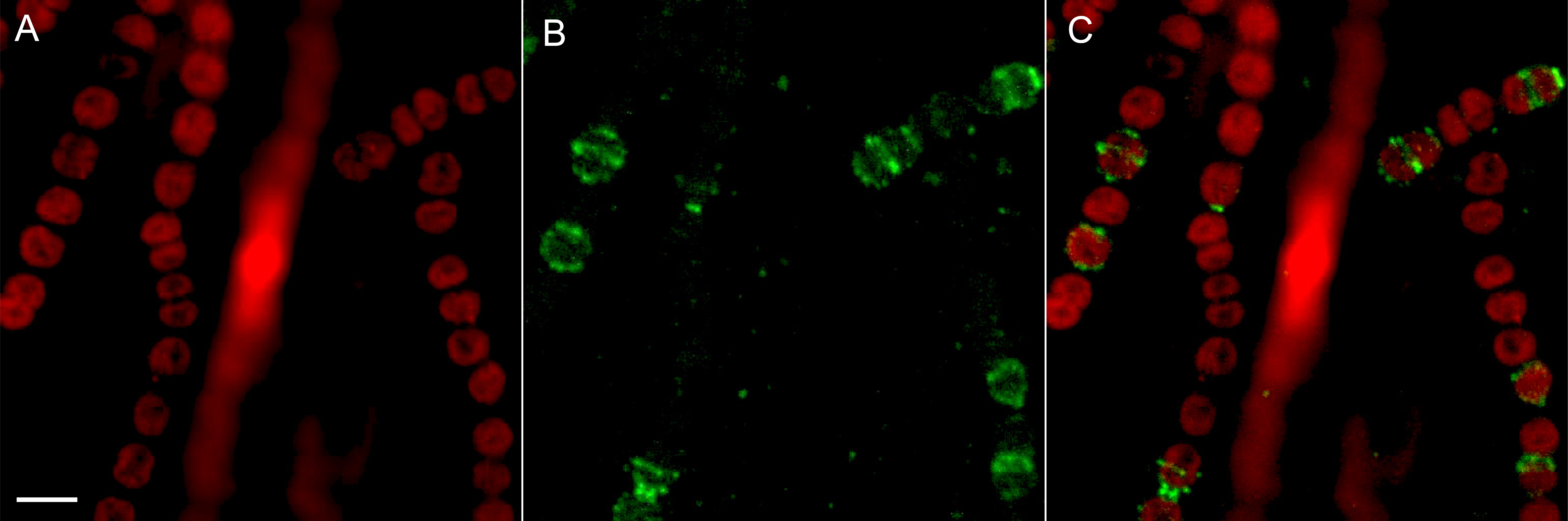
Figure 1. Immunolocalization of CyDiv in Anabaena sp. PCC7120. Deconvoluted image of a Z-stack. A. Autofluorescence; B. Image signal derived from primary antibody anti-CyDiv and secondary antibody Alexa Fluor 488 goat anti-rabbit IgG; C. Merged image of the autofluorescence and CyDiv-Alexa Fluor 488 fluorescence. White scale bar = 5 µm.
- Permeabilize the cells by adding a drop of 0.05% Triton X-100 in PBS for 2 min at room temperature, and repeat it three times by removing the drop each time with a pipette.
Data analysis
Images of Z-stacks were processed using ImageJ software (Schneider et al., 2012). For each channel of images, the point-spread function (PSF) was calculated using the Born and Wolf model within the PSF Generator plugin (Kirshner et al., 2013). Image deconvolution was performed with the Deconvolution Lab plugin with Richardson-Lucy algorithm using 10 iterations (Vonesch and Unser, 2008).
Recipes
- PBS buffer (pH 7.4)
137 mM NaCl
2.7 mM KCl
1.4 mM Na2HPO4
1.4 mM KH2PO4
Note: The PBS is filtered through a filter with a pore size of 0.2 µm and stored at room temperature.
Acknowledgments
The protocol described has been modified from (Plominsky et al., 2013; Miyagishima et al., 2014). This work was supported by Fondecyt grants #1131037, 1161232 and Fellowships for Graduate Student of Chilean Government # 21100780 and 21150983.
References
- Baulina, O. I. (2012). Ultrastructural plasticity of cyanobacteria. Springer Science & Business Media.
- Flores, E., Herrero, A., Forchhammer, K. and Maldener, I. (2016). Septal junctions in filamentous heterocyst-forming cyanobacteria. Trends Microbiol 24(2): 79-82.
- Jin, C., Mesqutia, M., Emelko, M., and Wong, A. (2016). Automated enumeration and size distribution analysis of Microcystis aeruginosa via fluorescence imaging. J Comput Vis Imaging Syst 2(1).
- Kehr, J. C. and Dittmann, E. (2015). Biosynthesis and function of extracellular glycans in cyanobacteria. Life (Basel) 5(1): 164-180.
- Kirshner, H., Aguet, F., Sage, D. and Unser, M. (2013). 3-D PSF fitting for fluorescence microscopy: implementation and localization application. J Microsc 249(1): 13-25.
- Mandakovic, D., Trigo, C., Andrade, D., Riquelme, B., Gomez-Lillo, G., Soto-Liebe, K., Diez, B. and Vasquez, M. (2016). CyDiv, a conserved and novel filamentous cyanobacterial cell division protein involved in septum localization. Front Microbiol 7: 94.
- Miyagishima, S. Y., Kabeya, Y., Sugita, C., Sugita, M. and Fujiwara, T. (2014). DipM is required for peptidoglycan hydrolysis during chloroplast division. BMC plant biology 14(1): 57.
- Plominsky, Á. M., Larsson, J., Bergman, B., Delherbe, N., Osses, I. and Vásquez, M. (2013). Dinitrogen fixation is restricted to the terminal heterocysts in the invasive cyanobacterium Cylindrospermopsis raciborskii CS-505. PloS one 8(2): e51682.
- Rippka, R. (1988). Recognition and identification of cyanobacteria. Methods enzymol 167: 28-67.
- Santamaria-Gomez, J., Ochoa de Alda, J. A., Olmedo-Verd, E., Bru-Martinez, R. and Luque, I. (2016). Sub-cellular localization and complex formation by aminoacyl-tRNA synthetases in cyanobacteria: Evidence for interaction of membrane-anchored ValRS with ATP synthase. Front Microbiol 7: 857.
- Schneider, C. A., Rasband, W. S. and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7): 671-675.
- Vonesch, C. and Unser, M. (2008). A fast thresholded landweber algorithm for wavelet-regularized multidimensional deconvolution. IEEE Trans Image Process 17(4): 539-549.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Trigo, C., Andrade, D. and Vásquez, M. (2017). Protein Localization in the Cyanobacterium Anabaena sp. PCC7120 Using Immunofluorescence Labeling. Bio-protocol 7(11): e2318. DOI: 10.21769/BioProtoc.2318.
Category
Microbiology > Microbial cell biology > Cell imaging
Cell Biology > Cell imaging > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












