- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ex vivo Model of Human Aortic Valve Bacterial Colonization
Published: Vol 7, Iss 11, Jun 5, 2017 DOI: 10.21769/BioProtoc.2316 Views: 7860
Reviewed by: Valentine V TrotterJose Antonio Reyes-DariasAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Ex vivo Culture and Contractile Force Measurements of Non-human Primate Heart Slices
Christine M. Poch [...] Alessandra Moretti
Jul 5, 2023 1503 Views

Analysis of Vascular Permeability by a Modified Miles Assay
Hilda Vargas-Robles [...] Michael Schnoor
Apr 5, 2025 2552 Views

An Ex Vivo Lung Histoculture Model for Studying Pulmonary Infection and Immune Response With SARS-CoV-2 as an Example of RNA Virus
Elena V. Maryukhnich [...] Elena J. Vasilieva
Dec 20, 2025 751 Views
Abstract
The interaction of pathogens with host tissues is a key step towards successful colonization and establishment of an infection. During bacteremia, pathogens can virtually reach all organs in the human body (e.g., heart, kidney, spleen) but host immunity, blood flow and tissue integrity generally prevents bacterial colonization. Yet, patients with cardiac conditions (e.g., congenital heart disease, atherosclerosis, calcific aortic stenosis, prosthetic valve recipients) are at a higher risk of bacterial infection. This protocol was adapted from an established ex vivo porcine heart adhesion model and takes advantage of the availability of heart tissues obtained from patients that underwent aortic valve replacement surgery. In this protocol, fresh tissues are used to assess the direct interaction of bacterial pathogens associated with cardiovascular infections, such as the oral bacterium Streptococcus mutans, with human aortic valve tissues.
Keywords: Streptococcus mutansBackground
The oral pathogen Streptococcus mutans is considered the major etiological agent in dental caries and can also be associated with extra-oral infections such as infective endocarditis (IE) (Banas, 2004). IE is generally initiated by a lesion of the heart valve endothelium which leads to the formation a sterile thrombus mainly composed of platelets, inflammatory cells, fibrin and other extracellular matrix (ECM) proteins (e.g., collagen, laminin) (Que and Moreillon, 2011). Other cardiovascular malignancies, such as calcific stenosis and atherosclerosis, can also cause tissue damage leading to the exposure and remodeling of ECM proteins (Yetkin and Waltenberger, 2009). This environment then provides suitable targets for colonization by different pathogens capable of interacting with host components. Thus, the development of relevant tools and experimental models may allow us to understand better how pathogens interact with heart tissues. Based on a previous protocol established by Chuang-Smith et al., 2010 using aortic heart valves from pigs, we developed an ex vivo tissue adherence assay using human heart valves obtained from patients that underwent aortic valve replacement (Freires et al., 2016). While this model does not reproduce the immunological responses and other host factors associated with the disease, it provides a relatively inexpensive system to assess the capacity of a given organism to directly interact with human heart valve tissues. Furthermore, while this model requires a close collaboration with a cardiac surgery unit, this type of surgery (i.e., aortic valve replacement) is routinely performed at health science centers in developed countries (Yetkin and Waltenberger, 2009).
Materials and Reagents
- Sterile specimen containers (Fisher Scientific, catalog number: 16-320-730 )
- 12-well tissue culture plates (Corning, Falcon®, catalog number: 351143 )
- Sterile culture tubes (4 ml) (Fisher Scientific, catalog number: 14-956-3D )
- Microcentrifuge tubes (1.7 ml) (Fisher Scientific, catalog number: S348903 )
- Glass scintillation vials (20 ml) (Sigma-Aldrich, catalog number: Z253081 )
- Sterile culture tubes (15 ml) (Fisher Scientific, catalog number: 14-956-6D )
- Desired bacterial strain(s) (e.g., Streptococcus mutans OMZ175)
- Extirpated heart tissues
- EGM-MV Bullet Kit (Lonza, catalog number: CC-3125 )
- Gentamicin (Sigma-Aldrich, catalog number: G1397 )
- Brain heart infusion medium (BHI) (BD, BactoTM, catalog number: 237500 )
- Hank’s balanced salt solution (HBSS) (Thermo Fisher Scientific, GibcoTM, catalog number: 14025092 )
- Erythromycin (Sigma-Aldrich, catalog number: E5389 )
- Kanamycin (Sigma-Aldrich, catalog number: K1377 )
- Sodium chloride (NaCl) (Avantor® Performance Materials, J.T. Baker®, catalog number: 3628-01 )
- Potassium chloride (KCl) (Avantor® Performance Materials, J.T. Baker®, catalog number: 3045-01 )
- Sodium phosphate dibasic (Na2HPO4) (Avantor® Performance Materials, J.T. Baker®, catalog number: 3827-01 )
- Potassium dihydrogen phosphate (KH2PO4) (Avantor® Performance Materials, J.T. Baker®, catalog number: 3246-01 )
- Paraformaldehyde (Sigma-Aldrich, catalog number: P6148 )
- Glutaraldehyde (Sigma-Aldrich, catalog number: 340855 )
- Sodium cacodylate trihydrate (Sigma-Aldrich, catalog number: C0250 )
- Agar (Fisher Scientific, catalog number: BP1423 )
- Fetal bovine serum (FBS) (Sigma-Aldrich, catalog number: F0392 )
- Hydrocortisone
- Bovine brain extract
- Human recombinant epidermal growth factor
- 1x phosphate buffer solution (PBS) (see Recipes)
- Fixative solution (see Recipes)
Equipment
- Biosafety cabinet class 2 (Nuaire, model: Labgard ES Energy Saver Class II, Type A2 , catalog number: NU-425-600)
- 3 mm skin biopsy punch (Acuderm, catalog number: P325 )
- Stainless steel forceps (Sigma-Aldrich, catalog number: Z168696 )
- Centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: HeraeusTM MultifugeTM 1 S-R )
- Vortex
- Motorized pestle (Kimble Chase Life Science and Research Products, catalog number: 7495400000 )
- pH meter
- Rocker (Reliable Scientific, model: 55D )
- CO2 incubator (VWR; model: 2325 )
- Zeiss-Auriga focused ion beam field emission scanning electron microscope (FIB-FE-SEM)
- Gatan Erlangshen digital camera
Procedure
- Sample collection and preparation
- Aortic valve tissues that would otherwise be discarded should be aseptically removed from patients undergoing aortic valve replacement by a cardiac surgery specialist.
- Immediately after surgery, tissues should be placed in a sterile specimen container, kept on ice and immediately taken to the laboratory for processing.
- Tissue preparation
- In a biosafety cabinet, open the vial containing the heart tissue and assess the level of calcification, inflammation and damage prior to further processing. Ideally, valve sections should be obtained from smooth areas with no calcification, signs of inflammation or residual blood (Figure 1).
- With a biopsy punch, press firmly on the selected areas of the tissue to cut through and remove the section from the rest of the tissues (Figure 1).
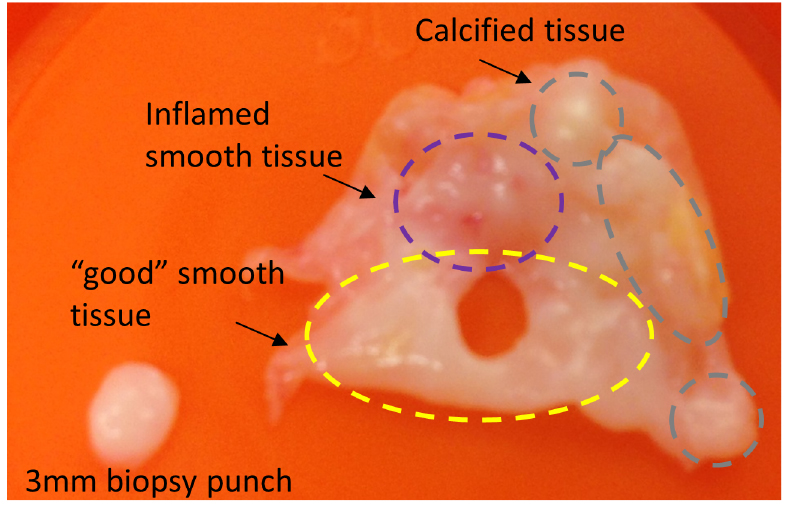
Figure 1. Selection of tissue from aortic valve tissues. After primary tissues are obtained, they should be immediately taken to the laboratory for evaluation and preparation. Only areas with no signs of calcification or inflammation are selected for sectioning with a 3 mm biopsy punch. - Use sterile forceps to collect the section and place it in individual wells of a 12-well plate containing 1 ml of supplemented EGM + gentamicin 300 µg ml-1.
- The number of sections that can be effectively obtained depends on the quality and the size of the tissue as well as on the type of experiment to be performed (see below).
- Incubate the 12-well plate containing the valve sections at 37 °C in a 5% CO2 atmosphere overnight (18-20 h).
- Bacterial attachment to aortic valve tissues
- On the same day of valve tissue processing, grow overnight bacterial cultures in triplicate. To grow S. mutans OMZ175, add 2 ml of BHI in a 4 ml sterile culture tube and inoculate by picking a single isolated colony from a freshly streaked plate.
- Incubate cultures at 37 °C in a 5% CO2 atmosphere overnight (18-20 h) without aeration.
- On the following day, wash the valve sections twice at room temperature with HBSS buffer to remove residual antibiotics. For this, add 1 ml of HBSS into two clean empty wells of the same plate and transfer the sections using sterile tweezers. Gently rock plate containing valve sections for 5 min and set the rocker to a setting of 40-50 motions per minute for each wash step.
- In the meantime, centrifuge the overnight cultures at 3,500 x g for 10 min at 4 °C, wash twice with sterile PBS and resuspend cultures with the initial culture volume.
- For each strain replicate, add 2.7 ml of fresh EGM without antibiotics to a clean 4 ml sterile culture tube and 300 µl of the bacterial culture making a 1:10 dilution of the bacterial suspension (approximately 1 x 107 CFU ml-1).
- Transfer washed valve sections to a new 12-well plate and add 1 ml of the bacterial suspension in EGM. In addition, take 100 µl of each replicate for serial dilution and plating of the initial culture CFU (T0).
- Incubate plate at 37 °C in a 5% CO2 atmosphere with gentle rocking for 90 min.
- After incubation, transfer each valve section into a 1.7 ml microcentrifuge tube containing 1 ml of HBSS and then vortex at medium intensity for 5 sec. This process should be performed three times in order to remove loosely bound bacteria.
- After washing, place each valve section in a 4 ml sterile culture tube containing 500 µl of PBS and homogenize the tissue for 2 min with a motorized pestle to detach tightly adhered bacteria.
- Take 100 µl aliquot from the homogenized valve suspension for serial dilution and plating of the final culture on BHI medium plates to determine the CFU (TF).
- Incubate inoculated plates at 37 °C in a 5% CO2 atmosphere. After 48 h, count colonies for CFU determination.
- Tissue preparation for analysis of adhesion by scanning electron microscopy (SEM)
- To visualize bacterial adhesion to heart valve sections by SEM, follow the inoculation protocol described above (steps 3a-3g).
- After incubation of tissue with bacteria, transfer each valve section into a 1.7 ml microcentrifuge tube containing 1 ml of HBSS and then vortex at medium intensity for 5 sec once.
Note: It is important not to perform too many washes at this stage as the SEM sample preparation includes several washing steps. - Upon washing, transfer the valve section to a labeled glass scintillation vial containing 15 ml of fixative solution.
- After the sample is fixed at 4 °C with gentle rocking for 48 h, it is ready for SEM analysis.
- Competition analysis of bacterial attachment
- On the same day of valve tissue processing, grow overnight bacterial cultures in triplicate. Unlike monoculture inoculations, this protocol requires marked strains so that they can be distinguished in CFU determination.
- For this study, S. mutans OMZ175 containing an erythromycin resistance cassette (ErmR) and S. mutans OMZ175 with the collagen binding protein gene (cnm) interrupted by a kanamycin resistance cassette (KanR) were used.
- To grow bacterial cultures, add 2 ml of BHI + Erm 10 µg ml-1 or BHI + Kan 1 mg ml-1 in a 4 ml sterile culture tube and inoculate by picking colonies from freshly streaked plates.
- Incubate cultures at 37 °C in a 5% CO2 atmosphere overnight (18-20 h) without aeration.
- On the following day, wash the valve sections twice at room temperature with HBSS buffer to remove residual antibiotics. For this, add 1 ml of HBSS into two clean empty wells of the same plate and transfer the sections using sterile tweezers. Gently rock plate containing valve sections for 5 min and set the rocker to a setting of 40-50 motions per minute for each wash step.
- In the meantime, centrifuge the overnight cultures at 3,500 x g for 10 min at 4 °C, wash twice with sterile PBS and resuspend cultures with the initial culture volume.
- For each strain replicate, add 2.7 ml of fresh EGM without antibiotics to a clean 4 ml sterile culture tube and 300 µl of the bacterial culture making a 1:10 dilution of the bacterial suspension (approximately 1 x 107 CFU ml-1).
- Transfer washed valve sections to a new 12-well plate and add 500 µl of the S. mutans OMZ175 (ErmR) and 500 µl of the S. mutans ∆cnm (KanR) bacterial suspensions in EGM. In addition, take 100 µl of each mixed bacterial replicate for serial dilution and plating of the initial culture CFU (T0).
- Incubate the inoculated valve sections at 37 °C in a 5% CO2 atmosphere with gentle rocking for 90 min.
- After incubation, transfer each valve section into a 1.7 ml microcentrifuge tube containing 1 ml of HBSS and then vortex at medium intensity for 5 sec. This process should be repeated three times in order to remove loosely bound bacteria.
- After washing, place each valve section in a 4 ml sterile culture tube containing 500 µl of PBS and homogenize the tissue for 2 min with a motorized pestle to detach tightly adhered bacteria.
- Take 100 µl aliquot from the homogenized valve suspension for serial dilution and plating of the final culture on both BHI + Erm 10 µg ml-1 and BHI + Kan 1 mg ml-1 plates to determine the CFU (TF).
- Incubate inoculated plates at 37 °C in a 5% CO2 atmosphere. After 48 h, count colonies for CFU determination.
Data analysis
- For visualization of bacterial adherence, tissues should be thoroughly examined by SEM (Figure 2). The top and bottom parts of the tissues, which are the areas of the heart valve continuously exposed to arterial flow and are generally undamaged, are referred to as smooth surfaces. The edge of tissues, which are displaying collagen fibers due to rupture by the biopsy punch, is referred to as rough surfaces.
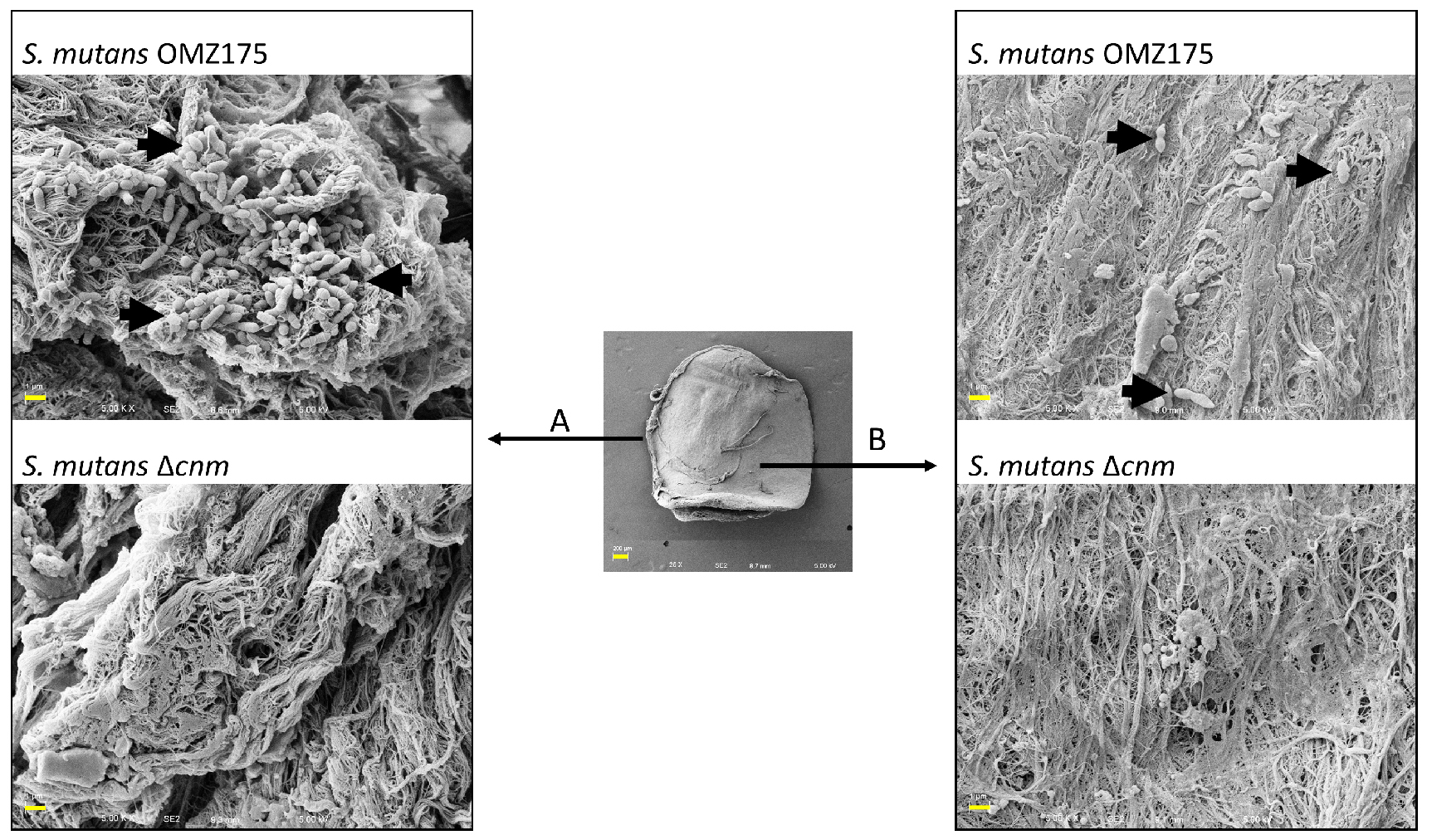
Figure 2. Adhesion of S. mutans OMZ175 to human aortic valve sections. Adhesion of S. mutans to human aortic valve sections analyzed by SEM. Center picture is a low magnification (25x) image of the valve section (scale bar = 200 µm). Representative images of attached bacteria to rough (A) and smooth surfaces (B) were taken at a 5,000x magnification (scale bars = 1 µm). The ex vivo adhesion of S. mutans OMZ175 (black arrows) to human valve sections requires the collagen-binding protein Cnm as no bacteria is detected in the ∆cnm strain. - To calculate the competitive index (CI), CFUs must be determined and applied in the following formula:
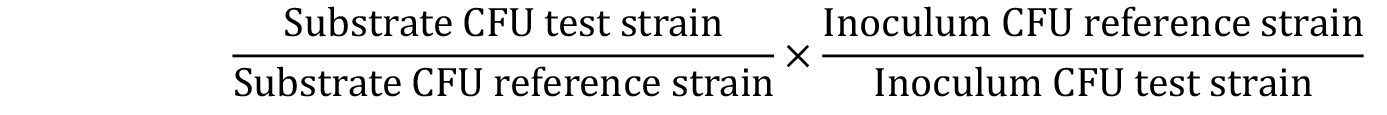
where, Inoculum = T0; Substrate = TF. - Values are then plotted on a column scatter plot and analyzed using one-way ANOVA with Bonferroni post hoc comparisons to determine substrate preferences among strain pairs. A CI value of 1 represents no difference in adherence between strains; a CI value lower than 1 represents less binding by test strain compared to the reference strain; a CI value higher than 1 represents more binding by the test strain compared to the reference strain.
A representative graph showing the competitive index (CI) of S. mutans OMZ175 and its ∆cnm counterpart is shown below. Each point in the graph represents an individual experiment using different tissue samples. The variability comes from the nature of the tissues since specimens are not exactly the same for every patient (Figure 3).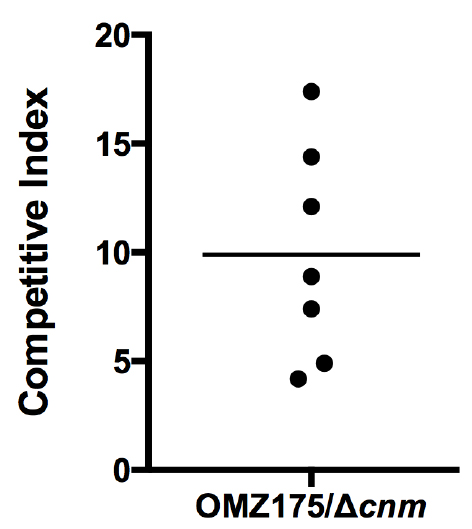
Figure 3. CI analysis of S. mutans OMZ175 and its ∆cnm counterpart. The CI values were calculated from CFU at T0 and TF for both strains using the CI formula described above. The mean CI was higher than 1 indicating that S. mutans OMZ175 outcompetes its ∆cnm counterpart. Expression of Cnm in OMZ175 enhances the bacterial capacity to adhere human aortic valve tissues.
Notes
- All protocols involving human subject must be previously approved by an Institutional Review Board. This protocol was established using discarded tissues from patients undergoing cardiac surgery for aortic valve replacement due to calcific stenosis. Patients younger than 18 years old, pregnant women and HIV-positive patients were excluded from the original study.
- All tissues should be individually and thoroughly evaluated prior to the ex vivo binding analysis. This is due to the different levels of calcification, inflammation and tissue damage in each patient.
- Tissues should be processed within 6 h of obtaining them. If required, tissues may be processed and placed in EGM + gentamicin 300 µg ml-1 for up to 72 h before performing colonization assay.
- For this study, tissue visualization by SEM was performed by the University of Rochester Medical Center Electron Microscopy Core. Samples were analyzed using a Zeiss-Auriga focused ion beam field emission scanning electron microscope (FIB-FE-SEM) and the images were captured using the attached Gatan Erlangshen digital camera.
- The number of input bacteria (total number of bacterial cells added to the tissues) for the competition experiment was previously determined to be approximately similar for S. mutans OMZ175 and its ∆cnm counterpart. Similarly, CFU input should be determined for any bacterial strains to be tested prior to competition analysis. To maintain reproducibility, we suggest adjusting bacterial cultures to specific OD600 nm to match the number of CFUs between strains.
Recipes
- 1x phosphate buffer solution (PBS)
137 mM NaCl
2.7 mM KCl
10 mM Na2HPO4
2 mM KH2PO4 - Fixative solution
4% paraformaldehyde
2.5% glutaraldehyde
0.1 M sodium cacodylate - EGM-MV Bulletkit
500 ml EGM complete medium
5% fetal bovine serum
500 mg hydrocortisone
6 mg bovine brain extract
0.1% human recombinant epidermal growth factor
Acknowledgments
We would like to thank Michael Swartz and Peter Knight from the University of Rochester for sample collection. This work was supported by the NIH-NIDCR (R01 DE022559). A.A.-R. was supported by NIH-NHLBI (F31 HL124951). I.A.F. was supported by the São Paulo Research Foundation (FAPESP, Brazil, 2013/25080-7). This protocol is a modified version from a study by Chuang-Smith et al., 2009.
References
- Banas, J. A. (2004). Virulence properties of Streptococcus mutans. Front Biosci 9: 1267-1277.
- Chuang-Smith, O. N., Wells, C. L., Henry-Stanley, M. J. and Dunny, G. M. (2010). Acceleration of Enterococcus faecalis biofilm formation by aggregation substance expression in an ex vivo model of cardiac valve colonization. PLoS One 5(12): e15798.
- Freires, I. A., Aviles-Reyes, A., Kitten, T., Simpson-Haidaris, P. J., Swartz, M., Knight, P. A., Rosalen, P. L., Lemos, J. A. and Abranches, J. (2016). Heterologous expression of Streptococcus mutans Cnm in Lactococcus lactis promotes intracellular invasion, adhesion to human cardiac tissues and virulence. Virulence 3: 1-12.
- Que, Y. A. and Moreillon, P. (2011). Infective endocarditis. Nat Rev Cardiol 8: 322-336.
- Yetkin, E. and Waltenberger, J. (2009). Molecular and cellular mechanisms of aortic stenosis. Int J Cardiol 135: 4-13.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Avilés-Reyes, A., Freires, I. A., Rosalen, P. L., Lemos, J. A. and Abranches, J. (2017). Ex vivo Model of Human Aortic Valve Bacterial Colonization. Bio-protocol 7(11): e2316. DOI: 10.21769/BioProtoc.2316.
Category
Microbiology > Microbe-host interactions > Ex vivo model
Cell Biology > Tissue analysis > Physiology
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












