- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Flow Cytometric Analysis of HIV-1 Transcriptional Activity in Response to shRNA Knockdown in A2 and A72 J-Lat Cell Lines
Published: Vol 7, Iss 11, Jun 5, 2017 DOI: 10.21769/BioProtoc.2314 Views: 11038
Reviewed by: Emilie BesnardEmilie BattivelliAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Epithelial and Stromal Cells from Colon Tissues in Homeostasis and Under Inflammatory Conditions
Clara Morral [...] Kathryn E. Hamilton
Sep 20, 2023 5706 Views

Isolation of Human Bone Marrow Non-hematopoietic Cells for Single-cell RNA Sequencing
Hongzhe Li [...] Stefan Scheding
Jun 20, 2024 2570 Views

Primary Neuronal Culture and Transient Transfection
Shun-Cheng Tseng [...] Eric Hwang
Jan 20, 2025 2729 Views
Abstract
The main obstacle to eradicating HIV-1 from patients is post-integration latency (Finzi et al., 1999). Antiretroviral treatments target only actively replicating virus, while latent infections that have low or no transcriptional activity remain untreated (Sedaghat et al., 2007). To eliminate viral reservoirs, one strategy focuses on reversing HIV-1 latency via ‘shock and kill’ (Deeks, 2012). The basis of this strategy is to overcome the molecular mechanisms of HIV-1 latency by therapeutically inducing viral gene and protein expression under antiretroviral therapy and to cause selective cell death via the lytic properties of the virus, or the immune system now recognizing the infected cells. Recently, a number of studies have described the therapeutic potential of pharmacologically inhibiting members of the bromodomain and extraterminal (BET) family of human bromodomain proteins (Filippakopoulos et al., 2010; Dawson et al., 2011; Delmore et al., 2011) that include BRD2, BRB3, BRD4 and BRDT. Small-molecule BET inhibitors, such as JQ1 (Filippakopoulos et al., 2010; Delmore et al., 2011), I-BET (Nicodeme et al., 2010), I-Bet151 (Dawson et al., 2011), and MS417 (Zhang et al., 2012) successfully activate HIV transcription and reverse viral latency in clonal cell lines and certain primary T-cell models of latency. To identify the mechanism by which BET proteins regulate HIV-1 latency, we utilized small hairpin RNAs (shRNAs) that target BRD2, BRD4 and Cyclin T1, which is a component of the critical HIV-1 cofactor positive transcription elongation factor b (P-TEFb) and interacts with BRD2, and tested them in the CD4+ J-Lat A2 and A72 cell lines. The following protocol describes a flow cytometry-based method to determine the amount of transcriptional activation of the HIV-1 LTR upon shRNA knockdown. This protocol is optimized for studying latently HIV-1-infected Jurkat (J-Lat) cell lines.
Keywords: Human immunodeficiency virus-1Background
A72 J-Lat cells contain a latent HIV minigenome composed of just the HIV promoter in the 5’LTR that drives the expression of the fluorescent marker GFP (LTR-GFP; A72) while in A2 cells transcriptional activity is driven by the viral transactivator Tat (LTR-Tat-IRES-GFP; A2) (Jordan et al., 2001 and 2003). HIV transcription can be induced in both cell lines with TNFα mimicking T cell-receptor engagement. Cells were transduced with lentiviral vectors expressing two different shRNAs targeting each cellular protein or a scrambled control, followed by puromycin treatment to select successfully transduced cells. Cells were then stimulated with a suboptimal or saturating dose of TNFα or were left unstimulated for 24 h, followed by flow cytometry of GFP to assess transcriptional activation of the HIV-1 LTR.
Materials and Reagents
- Production of shRNA containing virus particles
- 175 cm2 tissue culture flask (Corning, Falcon®, catalog number: 353112 )
- Tips
0.1-10 µl (Fisher Scientific, FisherbrandTM, catalog number: 02-681-440 )
1-200 µl (Fisher Scientific, FisherbrandTM, catalog number: 02-707-502 )
101-1,000 µl (Fisher Scientific, FisherbrandTM, catalog number: 02-707-509 ) - 15 ml conical tube (Fisher Scientific, FisherbrandTM, catalog number: 05-539-5 )
- 10 cm dish (Corning, catalog number: 353803 )
- 10 ml syringes (BD, catalog number: 309604 )
- 0.45 µm syringe filter (EMD Millipore, catalog number: SLHV033RS )
- HEK293T cells (ATCC, catalog number: CRL-3216 )
- shRNA-expressing lentiviral vectors (Sigma-Aldrich) to knockdown:
a.BRD2: TRCN0000006308 and TRCN0000006310
b.BRD4: TRCN0000021424 and TRCN0000021428
c.Cyclin T1: TRCN0000013673 and TRCN0000013675
d.Control: pLKO.1 vector containing scramble shRNA
e.Lentiviral packaging construct pCMVdelta R8.91 (Naldini et al., 1996)
f.VSV-G glycoprotein-expressing vector (Naldini et al., 1996) - DMEM (Mediatech, catalog number: 10-013-CV )
- Fetal bovine serum (FBS) (Gemini Bio-Products, catalog number: 100-106 )
- L-glutamine (Mediatech, catalog number: 25-005-Cl )
- 100x penicillin/streptomycin (Mediatech, catalog number: 30-002-Cl )
- 1x PBS (Mediatech, catalog number: 21-031-CV )
- Trypsin-EDTA (Mediatech, catalog number: 25-052-Cl )
- Chloroquine diphosphate salt (Sigma-Aldrich, catalog number: C6628 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 )
- Dextrose (Fisher Scientific, catalog number: BP350-1 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014 )
- Sodium phosphate dibasic (Na2HPO4) (Fisher Scientific, catalog number: BP332-500 )
- Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: C1016 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- Nuclease-free H2O (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9937 )
- Lenti-XTM p24 Rapid Titer Kit (Takara Bio, Clontech, catalog number: 632200 )
- 25 mM chloroquine (see Recipes)
- HBSS buffer (see Recipes)
- 2 M CaCl2 (see Recipes)
- Glycerol shock solution (see Recipes)
- 175 cm2 tissue culture flask (Corning, Falcon®, catalog number: 353112 )
- Infection of A2 and A72 J-Lat cells
- 75 cm2 tissue culture flask (Corning, Falcon®, catalog number: 353110 )
- 6 well tissue culture plates (Corning, Falcon®, catalog number: 353224 )
- Posi-ClickTM 1.7 ml microcentrifuge tubes (Denville Scientific, catalog number: C2170 )
- Tips
0.1-10 µl (Fisher Scientific, FisherbrandTM, catalog number: 02-681-440 )
1-200 µl (Fisher Scientific, FisherbrandTM, catalog number: 02-707-502 )
101-1,000 µl (Fisher Scientific, FisherbrandTM, catalog number: 02-707-509 ) - A2 and A72 J-Lat cells (Jordan et al., 2003)
- RPMI (Mediatech, catalog number: 10-040-CV )
- Fetal bovine serum (FBS) (Gemini Bio-Products, catalog number: 100-106 )
- L-glutamine (Mediatech, catalog number: 25-005-Cl )
- 100x penicillin/streptomycin (Mediatech, catalog number: 30-002-Cl )
- Polybrene® (Santa Cruz Biotechnology, catalog number: sc-134220 )
- Puromycin (Sigma-Aldrich, catalog number: P7255 )
- Polybrene solution (see Recipes)
- 75 cm2 tissue culture flask (Corning, Falcon®, catalog number: 353110 )
- Analysis of HIV-1 LTR transcriptional activation by flow cytometry
- 96-well tissue culture plates and lids (Thermo Fisher Scientific, Thermo ScientificTM, catalog numbers: 249570 and 163320 )
- Tips
0.1-10 µl (Fisher Scientific, FisherbrandTM, catalog number: 02-681-440 )
1-200 µl (Fisher Scientific, FisherbrandTM, catalog number: 02-707-502 )
101-1,000 µl (Fisher Scientific, FisherbrandTM, catalog number: 02-707-509 ) - A2 and A72 J-Lat cells (Jordan et al., 2003)
- 1x PBS (Mediatech, catalog number: 21-031-CV )
- RPMI (Mediatech, catalog number: 10-040-CV )
- Fetal bovine serum (FBS) (Gemini Bio-Products, catalog number: 100-106 )
- L-glutamine (Mediatech, catalog number: 25-005-Cl )
- 100x penicillin/streptomycin (Mediatech, catalog number: 30-002-Cl )
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418 )
- TNFα (PeproTech, catalog number: 300-01A ), make stock solution of 100 ng/μl in sterile H2O
- 7AAD, Propidium iodide or one of the Zombie viability dyes
- JQ1 (Cayman Chemical, catalog number: 11187 ), make stock solution of 10 mM in DMSO
- MACSQuant Running buffer (Miltenyi Biotech, catalog number: 130-092-747 )
- ApoTox-GloTM Triplex Assay (Promega, catalog number: G6320 )
- 96-well tissue culture plates and lids (Thermo Fisher Scientific, Thermo ScientificTM, catalog numbers: 249570 and 163320 )
- Confirmation of shRNA knockdown efficiency by SDS-PAGE and Western blot
- Posi-ClickTM 1.7 ml microcentrifuge tubes (Denville Scientific, catalog number: C2170 )
- Nitrocellulose membrane 0.2 µm (Bio-Rad Laboratories, catalog number: 1620112 )
- Whatman paper (GE Healthcare, catalog number: 3030-917 )
- High-performance chemiluminescence film (GE Healthcare, catalog number: 28906839 )
- Trizma® base (Sigma-Aldrich, catalog number: T1503 )
- NP-40 (Igepal CA-630) (Sigma-Aldrich, catalog number: I3021 )
- Na-deoxycholate (Deoxycholic acid, sodium salt) (Fisher Scientific, catalog number: BP349-100 )
- Sodium dodecyl sulfate (SDS) (Fisher Scientific, catalog number: BP166-500 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 )
- Ethylenediaminetetraacetic acid (EDTA) (Fisher Scientific, catalog number: S311-500 )
- Sodium fluoride (NaF) (Sigma-Aldrich, catalog number: S6521 )
Note: This product has been discontinued. - DCTM Protein assay (Reagents A, B and S, Bio-Rad Laboratories, catalog numbers: 5000113 , 5000114 , 5000115 )
- Mini-Protean® TGXTM Gels (Bio-Rad Laboratories, catalog number: 4569034 )
- PageRulerTM Prestained protein ladder (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 26617 )
- Antibodies for Western blot
- Rabbit polyclonal anti-BRD2 (Cell Signaling, catalog number: 5848 )
- Rabbit polyclonal anti-BRD4 (Abcam, catalog number: ab75898 )
- Rabbit polyclonal anti-Cyclin T1 (Santa Cruz Biotechnology, catalog number: sc-10750 )
- Rabbit polyclonal anti-α-Tubulin (Abcam, catalog number: ab15246 )
- Goat-anti-Rabbit IgG, HRP conjugated (Bethyl Laboratories, catalog number: A120-201P )
- Tween® 20 (Fisher Scientific, catalog number: BP337-500 )
- Glycine (Fisher Scientific, catalog number: BP381-5 )
- Methanol (Fisher Scientific, catalog number: A433P-4 )
- Blotting-grade blocker (dry milk) (Bio-Rad Laboratories, catalog number: 1706404 )
- Lumi-light Western blotting substrate (Roche Diagnostics, catalog number: 12015200001 )
- RIPA buffer (see Recipes)
- Western blot running buffer (see Recipes)
- Western blot transfer buffer (pH 8.3) (see Recipes)
- TBS-T buffer (see Recipes)
- Posi-ClickTM 1.7 ml microcentrifuge tubes (Denville Scientific, catalog number: C2170 )
Equipment
- Pipette
- Biosafety cabinet ‘Level 2’
- Tabletop centrifuge for Eppendorf tubes (Eppendorf, model: 5415 D )
- Tabletop centrifuge for 96-well plates, Eppendorf, 15 ml and 50 ml tubes; used for spininfection (Beckman Coulter, model: Allegra X-14R )
- CO2 tissue culture incubator, 37 °C (Thermo Electron, model: FormaTM Steri-CultTM CO2 Incubators, catalog number: 3307)
- MACSQuant VYB FACS analyzer (Miltenyi Biotech, model: MACSOuant® VYB , catalog number: 130-096-116)
- Mini-PROTEAN® Electrophoresis System (Bio-Rad Laboratories, catalog-numbers: 1658006FC and 1703935 )
- PowerPacTM HC (Bio-Rad Laboratories, model: Power Pac HC High-Current Power Supply , catalog number: 1645052)
- Rocker II (Boekel Scientific, catalog number: 260350 )
- SpectraMax MiniMaxTM 300 Imaging Cytometer (Molecular Devices, model: SpectraMax MiniMax 300 )
Software
- FlowJo 9.9 or never (Tree Star)
Procedure
- Production of shRNA containing virus particles in HEK293T cells
Note: HIV-1 particles are produced after calcium phosphate transfection in HEK293T cells.- HEK293T cell culture
- HEK293T cells are cultured in DMEM medium (supplemented with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin) in 175 cm2 tissue culture flasks in 35 ml medium.
- For maintenance of the HEK293T culture cells are, when approaching confluence (~80%), washed twice with PBS, then trypsinized (0.05% trypsin) and plated after 1/10 dilution in 175 cm2 tissue culture flasks. HEK293T cells are split every 2-3 days.
- One day before transfection, plate approximately 2 x 106 of HEK293T cells per 10 cm dish in 10 ml medium.
Note: After thawing frozen cell vials, HEK293T cells are cultured for at least one week before transfecting them for virus production. In order to maximize viral particle production, HEK293T cells are never kept more than 4 weeks in culture.
- HEK293T cells are cultured in DMEM medium (supplemented with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin) in 175 cm2 tissue culture flasks in 35 ml medium.
- Calcium phosphate transfection
Note: Protocol is described for transfection of one 10 cm dish. - Remove 2 ml medium from dish.
- Add 8 µl 25 mM chloroquine for a final concentration of 25 µM, incubate for about 5 min before adding the DNA/CaCl2 mix.
- Prepare DNA mix in nuclease-free H2O in a 15 ml conical tube (10 µg shRNA containing vector, 6.5 µg packaging construct pCMVdelta R8.91, and 3.5 µg VSV-G glycoprotein-expressing vector).
- Add nuclease-free H2O to the DNA mixture for a final volume of 876 µl.
- Add 1 ml of 2x HBSS buffer.
- Slowly (dropwise) add 124 μl of 2 M CaCl2 to the diluted DNA.
- After addition of CaCl2 gently vortex at the lowest setting for 5 sec (Note: Do not spin down!) and immediately add drop by drop the prepared DNA solution to the cells in the 10 cm dish.
- Swirl the plate to distribute the solution evenly.
- Culture for 3-4 h at 37 °C.
- Remove supernatant.
- Add 1 ml of glycerol shock solution, shock for one minute.
Note: The addition of glycerol results in an osmotic shock and is thought to cause the release of the DNA from the precipitate (Grosjean et al., 2006). - Aspirate shock solution, wash cells in 1 ml DMEM medium (supplemented with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin) and aspirate again.
- Add 7 ml of DMEM (supplemented with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin) medium.
- 48 h after transfection, collect supernatant with a 10 ml syringe and filter it through a sterile 0.45 µm PVDF membrane filter.
- Titer virus for p24 content using Lenti-XTM p24 Rapid Titer Kit according to manufacturer’s instructions.
- Filtered viral supernatant can be used fresh or stored at -80 °C.
- HEK293T cell culture
- Spin infection of A2 and A72 J-Lat cells with shRNA containing virus particles
Note: A2 and A72 cells are infected by spin infection.- A2 and A72 J-Lat suspension cell culture
- A2 and A72 cells are cultured in RPMI medium (supplemented with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin) in 75 cm2 tissue culture flasks in 40 ml medium. The flasks are placed horizontally to increase the surface area in which the cells are grown.
- Cells are split every 1-2 days and should be kept at a concentration of 2 x 105-1.5 x 106/ml.
- One day before transfection split A2 or A72 cells to 5 x 105/ml.
- A2 and A72 cells are cultured in RPMI medium (supplemented with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin) in 75 cm2 tissue culture flasks in 40 ml medium. The flasks are placed horizontally to increase the surface area in which the cells are grown.
- Infection of A2 and A72 J-Lat cells
- Count the cells and adjust cell number with new RPMI media (supplemented with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin). Add 1 x 106 of A2 or A72 cells to 1.7 ml microcentrifuge tubes.
- Centrifuge cell suspension for 3 min at 300 x g (1,800 rpm). Pipette off supernatant completely.
- Add 500 µl of shRNA virus (1 ng of p24 per 106 cells), 5 µl 1 M HEPES and 0.5 µl of polybrene (4 µg/ml) in 1.7 ml microcentrifuge tubes.
- Centrifuge at 32 °C for 1.5 h at 1,341 x g (2,400 rpm) (Tabletop centrifuge, Beckman).
- Pipette off supernatant completely.
- Resuspend cells in 3 ml RPMI medium (supplemented with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin) in 6-well tissue culture plates.
- Culture in CO2 incubator at 37 °C for one day.
- If shRNA-virus particles contain a puromycin resistant gene, select cells with 2 µg/ml RPMI medium of puromycin. RPMI media (supplemented with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin) with puromycin is changed every two days. Depending on how fast the cells grow and if they are needed for other experiments (e.g., chromatin immunoprecipitation, RNA isolation…) the cells can be grown out by adding media every 2 days and moved to bigger flasks or split to culture them in the 6-well plate.
- Culture in CO2 incubator at 37 °C for 4-10 days.
- Count the cells and adjust cell number with new RPMI media (supplemented with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin). Add 1 x 106 of A2 or A72 cells to 1.7 ml microcentrifuge tubes.
- A2 and A72 J-Lat suspension cell culture
- Analysis of HIV-1 LTR transcriptional activation by flow cytometry (please refer to the bio-protocol: Boehm and Ott, 2017)
- Count the cells and adjust cell number with new media. About 2 x 105/well of A2 or A72 cells are plated in 96-well tissue culture plates in 195 µl RPMI medium (supplemented with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin). For each condition perform three replicates.
- Cells are treated with 0.016-10 ng/ml of TNFα (as co-stimulus for maximal activation or to check for synergy), PBS or H2O control (depending on what was used as diluent for TNFα), or left untreated. TNFα and controls are pipetted in 5 µl to reach a total volume of 200 µl/well. Mix cells and TNFα by pipetting up and down at least 3 times.
- Culture in CO2 incubator at 37 °C for 18-48 h.
- Take the plate from the incubator and centrifuge at room temperature for 2 min at 1341 x g (2,400 rpm) (Tabletop centrifuge, Beckman).
- Then proceed directly to flow cytometry analysis. Cells do not have to be washed or fixed.
Notes:- For flow cytometry, MACSQuant VYB FACS analyzer was used to run 96-well plates. However, other flow cytometers such as Calibur or LSRII are also suitable for these experiments.
- If the analysis of cell viability by forward scatter (FSC) vs. side scatter (SSC) is not sufficient for accurate assessment of drug toxicity in drug studies, one of a variety of dyes/stains; for example 7AAD, Propidium iodide or one of the Zombie viability dyes can be used according to manufacturer’s instructions in the flow cytometric analysis in this protocol.
- For flow cytometry, MACSQuant VYB FACS analyzer was used to run 96-well plates. However, other flow cytometers such as Calibur or LSRII are also suitable for these experiments.
- Viability, cytotoxicity and apoptosis was measured with ApoTox-GloTM Triplex Assay (Promega) according to manufacturer’s instructions using a SpectraMax MiniMaxTM 300 Imaging Cytometer.
- Count the cells and adjust cell number with new media. About 2 x 105/well of A2 or A72 cells are plated in 96-well tissue culture plates in 195 µl RPMI medium (supplemented with 10% FBS, 1% L-glutamine and 1% penicillin/streptomycin). For each condition perform three replicates.
- Confirmation of shRNA knockdown efficiency by SDS-PAGE and Western blot
SDS-PAGE and Western blot protocols were described elsewhere (Renart et al., 1979; Towbin et al., 1979).- Lyse 1 x 106 cells in 100-500 µl RIPA buffer in 1.7 ml microcentrifuge tubes.
- Measure protein concentration by DCTM Protein Assay (Bio-Rad Laboratories) according to manufacturer’s instructions using a SpectraMax MiniMaxTM 300 Imaging Cytometer.
- Load PageRulerTM protein ladder in one lane and the other lanes equal amounts of protein from the cell lysates on 6-15% SDS gels (depending on the size of the protein) and electrophorese at 200 V for 1 h to separate the proteins. The expected size in kDa and the appropriate % of acrylamide for each protein are: BRD2, 110 kDa, 8%; BRD4, 171 kDa, 6.5%; Cyclin T1, 87 kDa, 8%; α-Tubulin, 50 kDa, 10%. Since the kDa of the proteins are so different the membranes can be cut to check for equal loading by incubation with α-Tubulin antibody.
- Transfer the separated proteins to the nitrocellulose membranes using a Bio-Rad Mini-PROTEAN® Electrophoresis System at 100 mA for 1-3 h or at 35 mA overnight (for proteins larger than 120 kDa).
- Block the transferred proteins on the membranes in TBS-T containing 5% nonfat dry milk (blocking buffer) for 1 h at room temperature on a rocker.
- Incubate the membranes with primary antibodies diluted (BRD2 1:1,000; BRD4 1 µg/ml; Cyclin T1 1:1,000; α-Tubulin 1:2,000) in TBS-T buffer over night at 4 °C on a rocker.
- Wash the membranes three times for approximately 5 min each time with TBS-T buffer and incubate with the HRP-conjugated secondary antibody, diluted 1:10,000 in TBS-T buffer, for 1 h at room temperature on a rocker.
- Wash the membranes three times for approximately 5 min each time with TBS-T buffer and incubate for 30 sec in Lumi-light Western blotting substrate.
- Develop on chemiluminescence films, expose the films for 10 sec up to 30 min. Depending on if the antibody has been reused and how many times it has been reused the signal will appear weaker, but cleaner (fewer background bands).
- Lyse 1 x 106 cells in 100-500 µl RIPA buffer in 1.7 ml microcentrifuge tubes.
Data analysis
Analysis of HIV-1 LTR transcriptional activation by flow cytometry (Figure 1).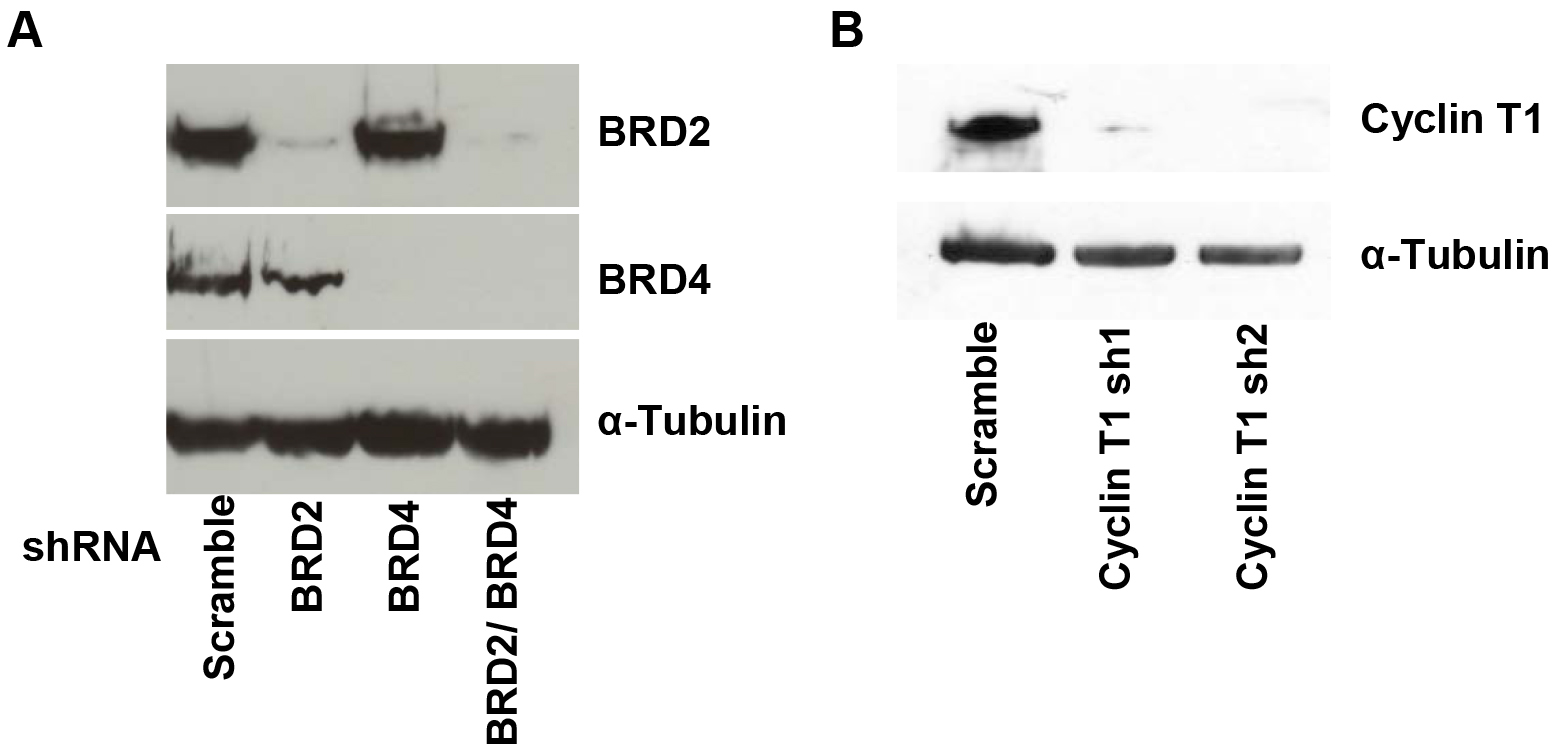
Figure 1. Western blot of BRD2, BRD4, Cyclin T1 and α-Tubulin. A. A72 cells were infected with virus containing shRNA constructs targeting BRD2, BRD4 or a nontargeting scramble control. At 4 days after infection, cells were harvested and lysed in Ripa buffer. Knockdown of BRD2 and BRD4 protein levels were confirmed by immunoblotting with BRD2 and BRD4 antibodies or the control α-Tubulin. B. A72 cells were infected with virus containing shRNA constructs targeting cyclin T1 or a nontargeting control. Knockdown of cyclin T1 protein levels are shown by immunoblotting with cyclin T1 or the control α-Tubulin antibody.
- First, set the gate on live J-Lat cells. Cell viability is monitored by forward (FSC-Area) and side scatter (SSC-Area) analysis (Figure 2A).
- Gate on singlets (FSC-Height vs. FSC-Area) (Figure 2B).
- Set the gate on SSC-Area and GFP/FITC-Area to identify the number of GFP+ cells (Figure 2C).
- Each sample is usually analyzed in triplicate and the experiment is independently repeated three times. Three replicates are averaged by calculating (GFP+ cells Experiment 1 + GFP+ cells Experiment 2 + GFP+ cells Experiment 3)/3. Also calculate standard deviation (STDEV) for error bars.
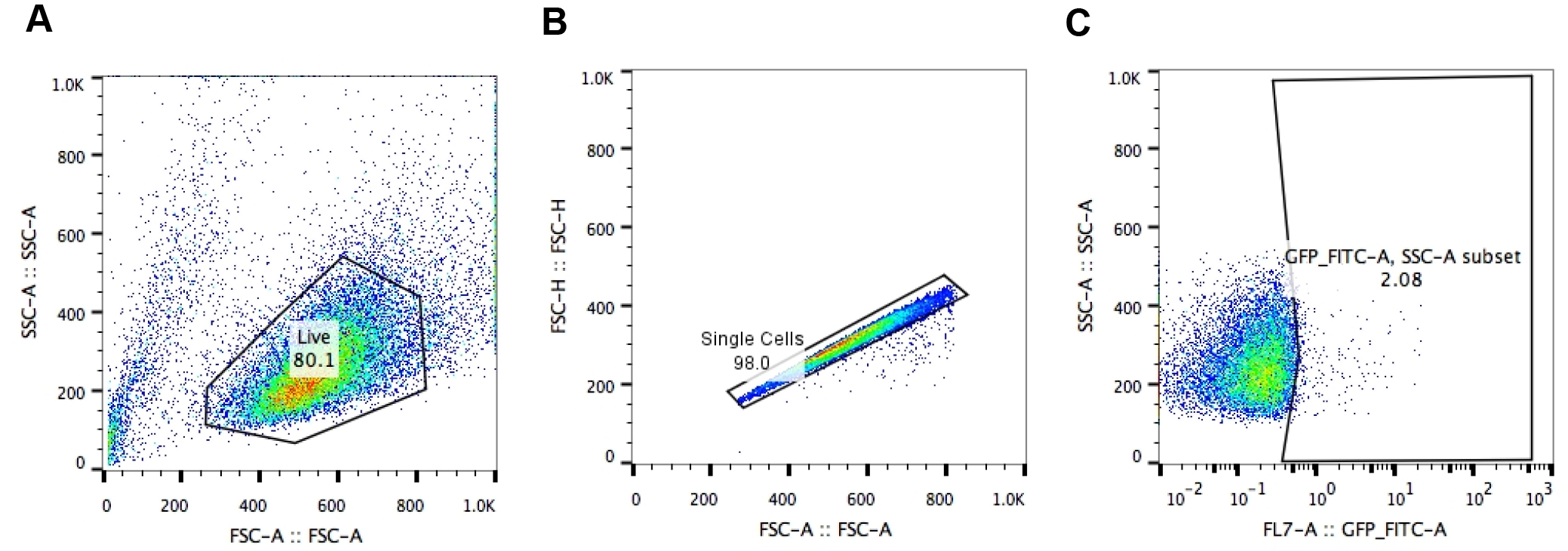
Figure 2. Analysis of HIV-1 LTR transcriptional activation by flow cytometry. Gating strategy to analyze A2 or A72 J-Lat cells: A. gating on live J-Lat cells based on size (FSC-Area) and granularity (SSC-Area); B. singlets gate (FSC-Height vs. FSC-Area); C. gating on GFP+ J-Lat cells (SSC-Area vs. GFP-FITC-Area).
Recipes
- 25 mM chloroquine
25 mM chloroquine in PBS - 2x HBSS
50 mM HEPES
10 mM KCl
12 mM dextrose
280 mM NaCl
1.5 mM Na2HPO4
Adjust pH to 7.1
Note: pH is crucial! - 2 M CaCl2
2 M CaCl2 in nuclease-free H2O - Glycerol shock solution
150 nM NaCl
20 mM HEPES, pH 7.0
15% glycerol - Polybrene solution
4 µg/ml in nuclease-free H2O - RIPA buffer
50 mM Tris buffer, pH 7.4
1% NP-40
0.5% Na-deoxycholate
0.1% SDS
150 mM NaCl
2 mM EDTA
50 mM sodium fluoride - Western blot running buffer
25 mM Trizma® base
190 mM glycine
0.1% SDS - Western blot transfer buffer (pH 8.3)
25 mM Trizma® base
190 mM glycine
20% methanol - TBS-T buffer
19 mM Trizma® base
2.7 mM potassium chloride
137 mM sodium chloride
1 ml Tween® 20
Acknowledgments
We thank Marielle Cavrois and the Flow cytometry core for the service provided for flow cytometry. This publication was made possible with the help from the University of California, San Francisco – Gladstone Institute of Virology & Immunology Center for AIDS Research (CFAR), a NIH-funded program (P30 AI027763). This research was supported as part of the amfAR Institute for HIV Cure Research, with funding from amfAR grant number 109301. Further, we gratefully acknowledge support from the California HIV/AIDS Research Program (Award number: F13-GI-316) to D.B., an industry-sponsored collaboration with JT Pharma that enabled the shRNA screen to M.O., and grant support from the CARE Collaboratory (U19 AI096113) and the NIH (RO1 AI083139 and RO1 DA043142) to M.O. This protocol was adapted from previous work: ‘BET bromodomain-targeting compounds reactivate HIV from latency via a Tat-independent mechanism’ (Boehm et al., 2013)
References
- Boehm, D., Calvanese, V., Dar, R. D., Xing, S., Schroeder, S., Martins, L., Aull, K., Li, P. C., Planelles, V., Bradner, J. E., Zhou, M. M., Siliciano, R. F., Weinberger, L., Verdin, E. and Ott, M. (2013). BET bromodomain-targeting compounds reactivate HIV from latency via a Tat-independent mechanism. Cell Cycle 12(3): 452-462.
- Boehm, D. and Ott, M. (2017). Flow cytometric analysis of drug-induced HIV-1 transcriptional activity in A2 and A72 J-Lat cell lines. Bio-protocol 7(10): e2290.
- Dawson, M. A., Prinjha, R. K., Dittmann, A., Giotopoulos, G., Bantscheff, M., Chan, W. I., Robson, S. C., Chung, C. W., Hopf, C., Savitski, M. M., Huthmacher, C., Gudgin, E., Lugo, D., Beinke, S., Chapman, T. D., Roberts, E. J., Soden, P. E., Auger, K. R., Mirguet, O., Doehner, K., Delwel, R., Burnett, A. K., Jeffrey, P., Drewes, G., Lee, K., Huntly, B. J. and Kouzarides, T. (2011). Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478(7370): 529-533.
- Deeks, S. G. (2012). HIV: Shock and kill. Nature 487(7408): 439-440.
- Delmore, J. E., Issa, G. C., Lemieux, M. E., Rahl, P. B., Shi, J., Jacobs, H. M., Kastritis, E., Gilpatrick, T., Paranal, R. M., Qi, J., Chesi, M., Schinzel, A. C., McKeown, M. R., Heffernan, T. P., Vakoc, C. R., Bergsagel, P. L., Ghobrial, I. M., Richardson, P. G., Young, R. A., Hahn, W. C., Anderson, K. C., Kung, A. L., Bradner, J. E. and Mitsiades, C. S. (2011). BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146(6): 904-917.
- Filippakopoulos, P., Qi, J., Picaud, S., Shen, Y., Smith, W. B., Fedorov, O., Morse, E. M., Keates, T., Hickman, T. T., Felletar, I., Philpott, M., Munro, S., McKeown, M. R., Wang, Y., Christie, A. L., West, N., Cameron, M. J., Schwartz, B., Heightman, T. D., La Thangue, N., French, C. A., Wiest, O., Kung, A. L., Knapp, S. and Bradner, J. E. (2010). Selective inhibition of BET bromodomains. Nature 468(7327): 1067-1073.
- Finzi, D., Blankson, J., Siliciano, J. D., Margolick, J. B., Chadwick, K., Pierson, T., Smith, K., Lisziewicz, J., Lori, F., Flexner, C., Quinn, T. C., Chaisson, R. E., Rosenberg, E., Walker, B., Gange, S., Gallant, J. and Siliciano, R. F. (1999). Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5(5): 512-517.
- Grosjean, F., Bertschinger, M., Hacker, D. L. and Wurm, F. M. (2006). Multiple glycerol shocks increase the calcium phosphate transfection of non-synchronized CHO cells. Biotechnol Lett 28(22): 1827-1833.
- Jordan, A., Bisgrove, D. and Verdin, E. (2003). HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22(8): 1868-1877.
- Jordan, A., Defechereux, P. and Verdin, E. (2001). The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J 20(7): 1726-1738.
- Naldini, L., Blomer, U., Gallay, P., Ory, D., Mulligan, R., Gage, F. H., Verma, I. M. and Trono, D. (1996). In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272(5259): 263-267.
- Nicodeme, E., Jeffrey, K. L., Schaefer, U., Beinke, S., Dewell, S., Chung, C. W., Chandwani, R., Marazzi, I., Wilson, P., Coste, H., White, J., Kirilovsky, J., Rice, C. M., Lora, J. M., Prinjha, R. K., Lee, K. and Tarakhovsky, A. (2010). Suppression of inflammation by a synthetic histone mimic. Nature 468(7327): 1119-1123.
- Renart, J., Reiser, J. and Stark, G. R. (1979). Transfer of proteins from gels to diazobenzyloxymethyl-paper and detection with antisera: a method for studying antibody specificity and antigen structure. Proc Natl Acad Sci U S A 76(7): 3116-3120.
- Sedaghat, A. R., Siliciano, J. D., Brennan, T. P., Wilke, C. O. and Siliciano, R. F. (2007). Limits on replenishment of the resting CD4+ T cell reservoir for HIV in patients on HAART. PLoS Pathog 3(8): e122.
- Towbin, H., Staehelin, T. and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 76(9): 4350-4354.
- Zhang, G., Liu, R., Zhong, Y., Plotnikov, A. N., Zhang, W., Zeng, L., Rusinova, E., Gerona-Nevarro, G., Moshkina, N., Joshua, J., Chuang, P. Y., Ohlmeyer, M., He, J. C. and Zhou, M. M. (2012). Down-regulation of NF-kappaB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J Biol Chem 287(34): 28840-28851.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Boehm, D. and Ott, M. (2017). Flow Cytometric Analysis of HIV-1 Transcriptional Activity in Response to shRNA Knockdown in A2 and A72 J-Lat Cell Lines. Bio-protocol 7(11): e2314. DOI: 10.21769/BioProtoc.2314.
Category
Cell Biology > Single cell analysis > Flow cytometry
Molecular Biology > DNA > Transfection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









