- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation and Infection of Drosophila Primary Hemocytes
Published: Vol 7, Iss 11, Jun 5, 2017 DOI: 10.21769/BioProtoc.2300 Views: 11202
Reviewed by: Ruth A. FranklinBenoit ChassaingRamalingam Bethunaickan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Bacterial Pathogen-mediated Suppression of Host Trafficking to Lysosomes: Fluorescence Microscopy-based DQ-Red BSA Analysis
Mădălina Mocăniță [...] Vanessa M. D'Costa
Mar 5, 2024 2902 Views

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2095 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1671 Views
Abstract
Phagocytosis of invading pathogens and their subsequent clearance in lysosomes is important for organismal fitness. We have devised the following protocol to extract phagocytic hemocytes from wild-type and mutant Drosophila larvae and infect the isolated hemocytes with GFP-labeled E. coli to measure the rate of phagocytosis and degradation within individual hemocytes over time.
Keywords: DrosophilaBackground
The experiment described below can be used to study phagosome biogenesis, maturation, and delivery to lysosomes. Bacterial accumulation has been well studied in the context of immuno-compromised Drosophila with defects in IMD or Toll signaling and the resulting reduced expression of antimicrobial peptides (e.g., Lemaitre and Hoffmann, 2007; Kleino and Silverman, 2014). Cellular responses to bacterial infections have been less investigated in Drosophila, with most studies focused on mutations that interfere with the initial phagocytic uptake of bacteria by hemocytes (Kocks et al., 2005; Parsons and Foley, 2016). Such bacterial uptake is straightforward to measure using FACS analysis (Tirouvanziam et al., 2004). For a detailed analysis of phagosomal maturation, however, we found it advantageous to examine individual hemocytes attached to a glass cover slip (Akbar et al., 2011; Rahman et al., 2012; Akbar et al., 2016) as this procedure offered us the best combination of temporal and spatial resolution for our studies of phagosome maturation.
Materials and Reagents
- Falcon tubes (15 ml) (Corning, Falcon®, catalog number: 352196 )
- Eppendorf tubes
- Cover glass, No 1.5, 22 mm2 (Corning, catalog number: 2850-22 )
- Petri dish (100 x 15 mm) (Corning, catalog number: 351029 )
- Kimwipe
- Micro slides (Corning, catalog number: 2948-75X25 )
- Sterile filter unit: 0.22 µm cellulose acetate filter flasks (Corning, catalog number: 430769 )
- Drosophila melanogaster wandering third instar larvae (Ashburner, 1989)
- E. coli (DH5α) constitutively expressing GFP
- peGFP (https://www.addgene.org/vector-database/2485/)
- pET-GFP-C11 (http://www.addgene.org/30183/)
- peGFP (https://www.addgene.org/vector-database/2485/)
- Bucket of ice
- LB growth media (Fisher Scientific, catalog number: BP1425-500 )
- Schneider’s Drosophila medium (Thermo Fisher Scientific, GibcoTM, catalog number: 21720 ) with 10% FBS (Atlanta Biologicals, Advantage, catalog number: S11095 )
- S2 cell media
- PBSS = 0.3% Saponin (Sigma-Aldrich, catalog number: S7900 ) in PBS
- 10% NGS
- Phalloidin Alexa 594 (Thermo Fisher Scientific, InvitrogenTM, catalog number: A12381 )
- Vectashield (Vector Laboratories, catalog number: H-1000 )
- Clear nail polish
- Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 )
- Sodium phosphate dibasic heptahydrate (Na2HPO4·7H2O) (Sigma-Aldrich, catalog number: S9390 )
- Potassium phosphate monobasic (KH2PO4) (Fisher Scientific, catalog number: P285 )
- Hydrochloric acid (HCl)
- Sodium hydroxide (NaOH)
- Paraformaldehyde (Electron Microscopy Sciences, catalog number: 19208 )
- 10% normal goat serum (Jackson ImmunoResearch, catalog number: 005-000-121 ) in PBSS
- 10x PBS (see Recipes)
- 8% paraformaldehyde (see Recipes)
- Fixative: 4% paraformaldehyde in PBS (see Recipes)
Equipment
- Spectrophotometer (Molecular Devices, model: SpectraMax M2 )
- Low speed centrifuge for 15 ml Falcon tubes (Eppendorf, model: 5804 R )
- Dissecting stereomicroscope with Leica L2 cold light source (Leica Microsystems, model: Leica L2 )
- Fine dissecting forceps (Fine Scientific Tools)
- 37 °C incubator with shaking (Eppendorf, New BrunswickTM, model: Innova® 44 )
- 25 °C incubator (BioCold Environment, model: BC49-IN )
- Dissecting dish
- Confocal Microscope
Note: We use a Zeiss LSM510 (Zeiss, model: LSM510 ) with 63 x NA1.4 objective.
Software
- ImageJ (NIH)
- Prism (GraphPad)
Procedure
- Preparation of GFP-expressing E. coli
- Inoculate 50 ml LB broth with a GFP E. coli colony.
- Incubate overnight at 37 °C with shaking.
- Use spectrophotometer to measure optical density at 600 nm (OD600).
- Measure bacteria with broth, and broth alone.
- Subtract OD600 of fresh LB broth from bacteria with broth.
- Measure bacteria with broth, and broth alone.
- Dilute bacteria to OD600 = 0.1
- Spin 10 ml of bacterial solution at 4,000 x g for 10 min in 15 ml Falcon tubes at room temperature.
- Decant LB broth.
- Re-suspend, by vortexing in sufficient 1x PBS, to adjust bacteria to an OD600 of 0.1.
- Aliquot 1 ml into Eppendorf tubes.
- Heat-kill by placing tubes at 65 °C for 20 min.
- Freeze aliquots and thaw when ready to use.
- Re-suspend bacteria in Schneider’s Drosophila medium with FBS
- Centrifuge Eppendorf tube with heat-killed GFP-bacteria at 4,000 x g for 10 min at room temperature.
- Decant PBS supernatant.
- Add 1 ml Schneider’s Drosophila medium and vortex to re-suspend.
- For the E. coli we use this yields about 40,000 bacteria per µl of solution.
- Once re-suspended, warm to 25 °C before exposure to cells.
- Inoculate 50 ml LB broth with a GFP E. coli colony.
- Primary hemocyte isolation
- Warm fresh Schneider’s Drosophila medium to 25 °C.
- Collect ten wandering third instar larvae in a large drop of water on dissecting dish–wash thoroughly and place in fresh drop.
- Place a 22 mm2 coverslip in a 100 x 15 mm Petri dish.
- Add 200 μl Schneider’s Drosophila medium from step B1 to the coverslip.
- This should create a large droplet.
- Keep the area of the droplet as small as possible.
- Do not let the droplet spread out across the coverslip.
- This should create a large droplet.
- Place the washed larvae into the large droplet on the coverslip.
- Under a dissecting microscope, use forceps to pinch and immobilize the tail end of the larva and use another forceps to grab and rip the outer cuticle of the larvae from tail to mouth (see Figure 1).
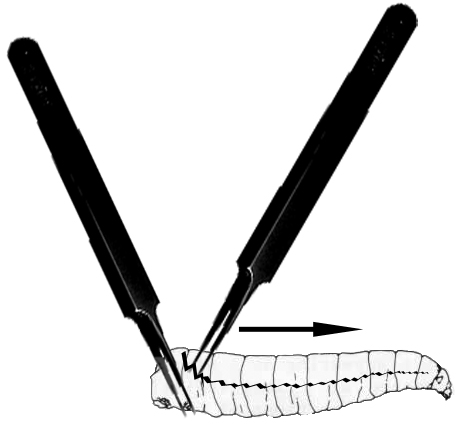
Figure 1. Larval dissection. This image depicts the way forceps are used to hold (left forceps) and dissect the cuticle of larvae covered with S2 cell media with the right forceps along the larval midline (jagged black line) thereby releasing the larval hemolymph into the S2 cell media. - Rip open all ten larvae in the media droplet on the coverslip.
Note: Do this step as quickly as possible. - Remove larval carcasses from the media droplet.
- Incubate hemolymph in media on coverslip in the Petri dish to allow hemocytes to attach to coverslip (at 25 °C for 15 min).
- This 15-min incubation will be sufficient time for hemocytes to adhere to the coverslip.
- After about 10 min, melanization may begin to occur (see Figure 2).
- Begin washes with PBS if the media features small black spots. These indicate the beginning of melanization that we seek to avoid.
- Do not allow the media to turn black.
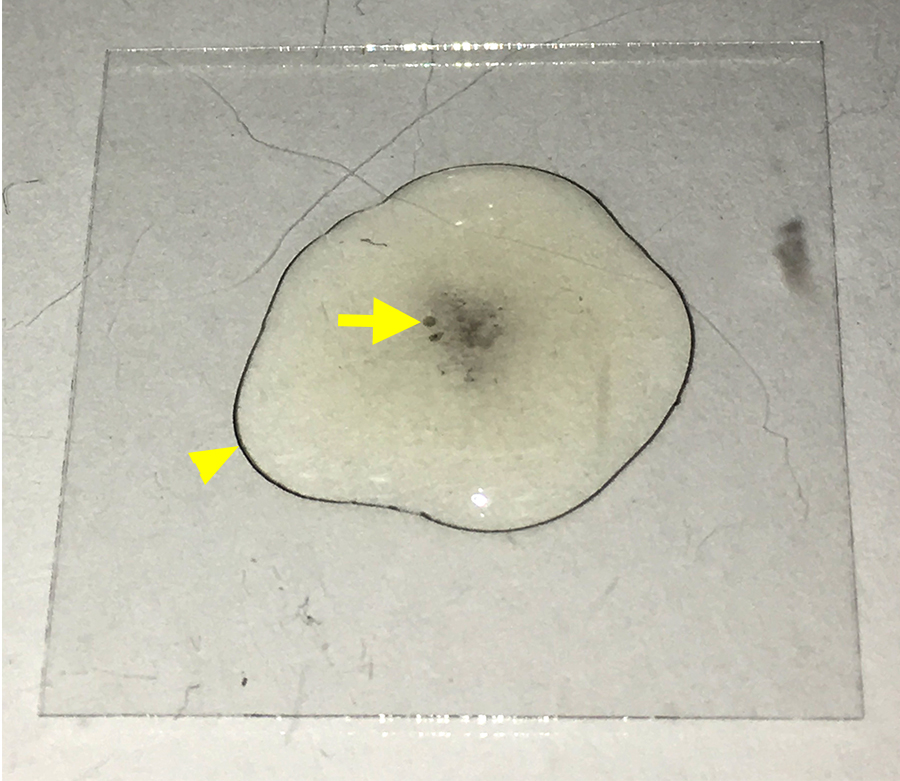
Figure 2. Early signs of melanization. The image displays a coverslip (22 x 22 mm) with a drop of medium containing hemocytes settling on the coverslip. Over time, crystal cells in the medium will initiate melanization that can be detected by the dark spots in the solution (arrow) and the dark ring at the edge of the drop (arrowhead). At this stage, hemocytes should be washed with PBS to remove crystal cells.
- This 15-min incubation will be sufficient time for hemocytes to adhere to the coverslip.
- Pour 1x PBS carefully into the Petri dish away from the coverslip to avoid dislodging the adherent cells.
- Wash thoroughly by slowly swirling the dish.
- Pour off PBS from Petri dish.
- Repeat wash (B10).
- Wash thoroughly by slowly swirling the dish.
- Hemocytes will adhere to the coverslip, while the PBS washes remove the majority of crystal cells, which are mainly responsible for melanization.
- Remove the coverslip with forceps and place on Kimwipe, then place in a dry Petri dish on ice.
- Warm fresh Schneider’s Drosophila medium to 25 °C.
- Expose hemocytes to GFP bacteria
- Add enough GFP bacteria in Schneider’s Drosophila medium from step A5 (40,000 bacteria/µl) to cover the entire coverslip without spilling over the edge of the coverslip (~200 µl).
- Incubate bacteria with hemocytes on ice for 20 min to allow the bacteria to adhere to the cell surface.
- Rinse 3 x by pouring PBS into the Petri dish and swirling gently. Remove PBS by pouring off.
- Remove coverslip with forceps and place on Kimwipe, then place in dry Petri dish.
- Add fresh, warm Schneider’s Drosophila medium on the coverslip for the desired amount of chase time (in wild-type hemocytes the majority of bacteria will be digested and undetectable by 45 min).
- Add enough GFP bacteria in Schneider’s Drosophila medium from step A5 (40,000 bacteria/µl) to cover the entire coverslip without spilling over the edge of the coverslip (~200 µl).
- Fix, stain, and mount hemocytes (all performed at room temperature)
- Wash the media with PBS as before.
- Add enough 4% paraformaldehyde to cover the coverslip without spilling over its edges.
- Fix cells for 30 min.
- Thoroughly wash cells with PBS as before.
- Add PBSS for 30 min to permeabilize cell membranes.
- Remove PBSS and add 10% NGS in PBSS for 30 min to block unspecific antibody binding.
- Stain with primary and secondary antibodies of your choice and/or Phalloidin-Alexa 594 in PBSS for one hour.
Note: Keep specimen in the dark as much as possible during and after secondary antibody staining. - Wash with PBS 3 x 15 min after each antibody.
- Place the coverslip on Kimwipe to dry it.
- Add a small drop of Vectashield to a microscope slide.
- Slowly lay the coverslip with stained hemocytes onto the drop of Vectashield.
- Seal the edges of the coverslip to the slide with nail polish.
- Image the cells on a confocal microscope with a 63x objective and 3x digital zoom.
- Wash the media with PBS as before.
Data analysis
To determine the number of remaining bacteria at different time points in different genetic backgrounds, confocal images were opened with ImageJ (NIH) and smoothed with a Gaussian blur of one before merging bacteria and phalloidin-stained channels. The number of bacteria within individual cells was counted and recorded in a Prism (GraphPad) spreadsheet as a single data point for each cell. Bacteria were counted when the entire GFP bacterium was surrounded by phalloidin staining. At least 25 cells were counted for each of three experiments. Prism software was used to plot a box and whisker graph and to perform a one-way ANOVA comparing relevant data sets (all to all).
Notes
- The experiment described above has been beneficial to our lab’s analysis of bacterial accumulation and phagosome maturation within primary hemocytes (Figure 3) and the effect of different genotypes (Akbar et al., 2011; Rahman et al., 2012; Akbar et al., 2016). Chase time and antibody conditions can be adapted for a wide variety of experimental questions. However, great care must be taken during the initial steps of hemocyte isolation from larvae. As mentioned above, the Schneider’s Drosophila medium cell media droplet must maintain a dome-like shape in order to contain the larvae within the droplet and to keep hemocytes in a defined area with ample nutrients.
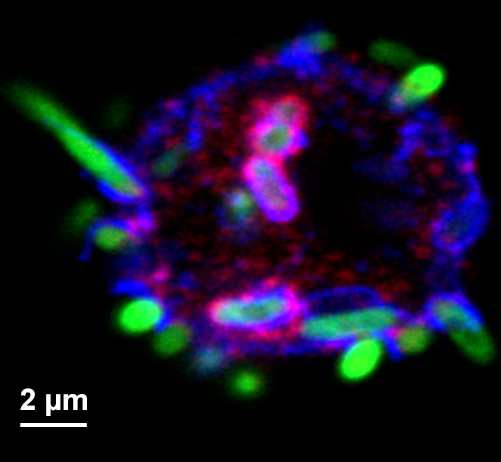
Figure 3. Micrograph of hemocyte and GFP-expressing bacteria of various stages of phagocytosis. Hemocyte surface, phagosomes and phagolysosomes are highlighted by staining for Hook (red, Krämer and Phistry, 1996) and Spinster (blue, Sweeney and Davis, 2002). - Hemocytes isolated by our procedure are mostly phagocytic active plasmatocytes, but also contain crystal cells that make up some 5% of the hemocyte pool (Tepass et al., 1994). Upon activation crystal cells secrete prophenoloxidase which is activated by proteolysis. Subsequently, the activated phenoloxidase initiates a melanization reaction that interferes with imaging of bacterial degradation products (Neyen et al., 2014). Therefore, after all the larvae have been ripped open and subsequently removed from the media, it is critically important to prevent the Schneider’s Drosophila medium from turning black. The melanization process will start around 10 min after the larval dissecting begins and will proceed with hemocytes cannibalizing other hemolymph cells (i.e., crystal cells). Other researchers (Tirouvanziam et al., 2004) added protease inhibitors to the isolation medium to suppress the protease cascade that initiates melanization. We found, however, that our approach was sufficient to minimize melanization, while avoiding any potential effect of the protease inhibitor on phagosome maturation and bacterial degradation.
Recipes
- 10x PBS stock solution
80 g NaCl
2 g KCl
26.8 g Na2HPO4·7H2O
2.4 g KH2PO4
Dissolve in 800 ml ddH2O
Adjust pH with HCl to 7.4
Add ddH2O to 1 L and mix well - 8% paraformaldehyde stock solution
- Heat 200 ml dH2O to 55 °C (but not warmer!)
- Add 2 drops 50% w/w NaOH
- Add 20 g paraformaldehyde
- Stir continuously until clear
- Vacuum filter through with 0.22 µm filter
- Add ddH2O to adjust final volume to 250 ml
- Aliquot 2 ml into 15 ml Falcon tubes
- store at -80 °C
- Fixative: 4% paraformaldehyde in PBS
- Thaw a 2 ml aliquot of 8% paraformaldehyde
- Add 1.6 ml dH2O
- Add 400 µl 10x PBS stock solution
- Mix well
Acknowledgments
The work herein was supported by NIH Grants EY010199, EY021922. This protocol has been adapted and modified from our previous published work (Akbar et al., 2011; Akbar et al., 2016). The authors of this work declare no conflicts of interest.
References
- Ashburner, M. (1989). Drosophila: A laboratory manual. Cold Spring Harbor Laboratory Press.
- Akbar, M. A., Mandraju, R., Tracy, C., Hu, W., Pasare, C. and Kramer, H. (2016). ARC syndrome-linked Vps33B protein is required for inflammatory endosomal maturation and signal termination. Immunity 45(2): 267-279.
- Akbar, M. A., Tracy, C., Kahr, W. H. and Kramer, H. (2011). The full-of-bacteria gene is required for phagosome maturation during immune defense in Drosophila. J Cell Biol 192(3): 383-390.
- Kleino, A. and Silverman, N. (2014). The Drosophila IMD pathway in the activation of the humoral immune response. Dev Comp Immunol 42(1): 25-35.
- Kocks, C., Cho, J. H., Nehme, N., Ulvila, J., Pearson, A. M., Meister, M., Strom, C., Conto, S. L., Hetru, C., Stuart, L. M., Stehle, T., Hoffmann, J. A., Reichhart, J. M., Ferrandon, D., Ramet, M. and Ezekowitz, R. A. (2005). Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell 123(2): 335-346.
- Krämer, H., and Phistry, M. (1996). Mutations in the Drosophila hook gene inhibit endocytosis of the boss transmembrane ligand into multivesicular bodies. J Cell Biol 133(6): 1205-1215.
- Lemaitre, B. and Hoffmann, J. (2007). The host defense of Drosophila melanogaster. Annu Rev Immunol 25: 697-743.
- Neyen, C., Bretscher, A. J., Binggeli, O. and Lemaitre, B. (2014). Methods to study Drosophila immunity. Methods 68(1): 116-128.
- Parsons, B. and Foley, E. (2016). Cellular immune defenses of Drosophila melanogaster. Dev Comp Immunol 58: 95-101.
- Rahman, M., Haberman, A., Tracy, C., Ray, S. and Kramer, H. (2012). Drosophila mauve mutants reveal a role of LYST homologs late in the maturation of phagosomes and autophagosomes. Traffic 13(12): 1680-1692.
- Sweeney, S.T., and Davis, G.W. (2002). Unrestricted synaptic growth in spinster-a late endosomal protein implicated in TGF-beta-mediated synaptic growth regulation. Neuron 36(3): 403-416.
- Tepass, U., Fessler, L. I., Aziz, A. and Hartenstein, V. (1994). Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development 120(7): 1829-1837.
- Tirouvanziam, R., Davidson, C. J., Lipsick, J. S. and Herzenberg, L. A. (2004). Fluorescence-activated cell sorting (FACS) of Drosophila hemocytes reveals important functional similarities to mammalian leukocytes. Proc Natl Acad Sci U S A 101(9): 2912-2917.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tracy, C. and Krämer, H. (2017). Isolation and Infection of Drosophila Primary Hemocytes. Bio-protocol 7(11): e2300. DOI: 10.21769/BioProtoc.2300.
Category
Immunology > Animal model > Drosophila (Fruit fly)
Microbiology > Microbe-host interactions > Bacterium
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










