- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Imaging the Pharynx to Measure the Uptake of Doxorubicin in Caenorhabditis elegans
Published: Vol 7, Iss 10, May 20, 2017 DOI: 10.21769/BioProtoc.2291 Views: 8679
Reviewed by: Neelanjan BoseKanika GeraJianwei Sun

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Labelling of Active Transcription Sites with Argonaute NRDE-3—Image Active Transcription Sites in vivo in Caenorhabditis elegans
Antoine Barrière and Vincent Bertrand
Jun 5, 2022 2689 Views

Monitoring the Recruitment and Fusion of Autophagosomes to Phagosomes During the Clearance of Apoptotic Cells in the Nematode Caenorhabditis elegans
Omar Peña-Ramos and Zheng Zhou
Nov 20, 2022 2137 Views

Preparation of Caenorhabditis elegans for Scoring of Muscle-derived Exophers
Katarzyna Banasiak [...] Wojciech Pokrzywa
Jan 5, 2023 1882 Views
Abstract
Caenorhabditis elegans offers an array of advantages to investigate the roles of uptake transporters. Herein, an epifluorescent microscopy approach was developed to monitor the uptake of the autofluorescent anticancer drug, doxorubicin, into the pharynx of C. elegans by organic cation transporters.
Keywords: C. elegansBackground
Human cells have over 450 solute carrier transporters that are believed to facilitate the uptake of several ions, nutrients, as well as both therapeutic and anticancer drugs (Cesar-Razquin et al., 2015). However, the roles and substrates of a large number of these uptake transporters are not known. C. elegans is an inexpensive model organism that offers a multitude of advantages over mammalian cells to rapidly study many biological processes that are highly conserved in nature. During the last decade, this organism has been instrumental in several drug discovery programs to identify novel small molecules, e.g., those that act as antimicrobials and inhibit oxidative stress, although the yield of bioactive compounds has been less striking (Burns et al., 2010; O’Reilly et al., 2014). We reason that the recovery rate could be higher if there is greater and selective influx of the molecules by uptake transporters into the animal cells. To date, only three studies have been performed to understand the roles of uptake transporters in C. elegans (Wu et al., 1999; Cheah et al., 2013; Papaluca and Ramotar, 2016). Thus, characterization of the function and substrate specificities of uptake transporters in this organism will be advantageous towards improving the strategies employed to identify novel bioactive molecules. Herein, we outline a method to monitor uptake of the anticancer drug doxorubicin into the pharynx of C. elegans (Papaluca and Ramotar, 2016). Doxorubicin autofluorescence can be readily monitored by several widely available detection systems such as the epifluorescent microscope. We note that several benefits can be derived from this approach including a hunt for novel therapeutic substrates of the transporter by competing for doxorubicin uptake.
Materials and Reagents
- Petri dish 60 x 15 mm (SARSTEDT, catalog number: 82.1194.500 )
- 15 ml conical tube
- Frosted microscope slides (size: 1 x 3” ; thickness: 1-2 mm) (UltiDent Scientific, catalog number: 170-7107A )
- Microscope cover glass (size: 22 x 22 #1.5) (Fisher Scientific, catalog number: 12-541B )
- Platinum wire (Thomas Scientific, catalog number: 1233S71 )
- Pasteur pipet (Fisher Scientific, catalog number: 13-678-20C )
- E. coli bacteria HT115DE3 with the plasmid pL4440-empty vector
- E. coli bacteria HT115DE3 with the plasmid pL4440-oct-1 (Ahringer’s collection). Sequence verified
- E. coli bacteria HT115DE3 with the plasmid pL4440-oct-2 (Ahringer’s collection). Sequence verified
- Bristol N2 (wild type) and RB1084 [oct-1(ok1051) I] from Caenorhabditis Genetic Center
- Ampicillin (Sigma-Aldrich, catalog number: A9518 )
- Potassium phosphate monobasic (KH2PO4) (Bio Basic, catalog number: PB0445 )
- Magnesium sulfate (MgSO4) (Bio Basic, catalog number: MRB0329 )
- Calcium chloride dihydrate (CaCl2) (Fisher Scientific, catalog number: C79-500 )
- Cholesterol (Sigma-Aldrich, catalog number: C8503 )
- Ethanol 100% (works from any company)
- Doxorubicin (for Research from Hôpital Maisonneuve-Rosemont, Montreal, Canada). Stock concentration at 2 mg/ml
- IPTG (Bio Basic, catalog number: PRB0447 )
- Levamisol hydrochloride (MP Biomedical, catalog number: 155228 )
- Clear nail polish from Wild Shine from Dollarama
- Tryptone (Bio Basic, catalog number: TG217 (G211)) for Luria Broth (LB) media
- Yeast extract (Wisent Bioproducts, catalog number: 800-150-LG ) for LB media
- Sodium chloride (NaCl) (Wisent Bioproducts, catalog number: 600-082 )
- Bacteriological agar (Wisent Bioproducts, catalog number: 800-010-CG )
- Peptone (Wisent Bioproducts, catalog number: 800-157-LG ) for nematode grown media (NGM)
- Agarose (Wisent Bioproducts, catalog number: 800-015-CG )
- Sodium hydroxide (NaOH) (Bio Basic, catalog number: SB0617 )
- Bleach Lavo Pro6 (Lavo Inc, Montreal, Canada)
- Sodium phosphate dibasic (Na2HPO4) (Bio Basic, catalog number: S0404 )
- LB solution (see Recipes)
- Nematode growth media (NGM) (see Recipes)
- Agar pad (see Recipes)
- Alkaline Hypochlorite solution for bleaching the worms (see Recipes)
- M9 buffer (see Recipes)
Equipment
- Incubator at 20 °C, but with a range from 15 to 37 °C (SHEL LAB, model: 2020 )
- 37 °C incubator (Panasonic Healthcare, model: Mir-262 )
- 37 °C shaker (Inforst, model: Multitron Standard )
- 500 ml glass bottle (Wheaton graduated glass media bottles with lined caps)
- Microwave (inverter model, Panasonic Healthcare)
- 55 °C water bath (Precision Scientific, catalog number: 66800 )
- Metal spatula (VWR)
- Flame
- Neutrex culture tubes 16 x 15 mm
- Pyrex Erlenmeyer flask different sizes for bacteria culturing
- Autoclave
- Stereomicroscope Leica MZ 8 (Leica 10445538 Plan Microscope Objective Lens 1.0x) (Leica, model: MZ 8 )
- DeltaVision Elite Restoration System (GE Healthcare, model: DeltaVision Elite High Resolution Microscope ) and the DeltaVision imaging system user’s manual
- Fisher Vortex Genie 2 (Fisher Scientific, catalog number: 12-812 )
- Eppendorf 5810 R centrifuge (Eppendorf, model: 5810 R )
- VWR rocking platform shakers basic (VWR)
- Ptc-100 Programmable Thermal Controller 96 Well (Bio-Rad Laboratories, model: Ptc-100® Programmable Thermal Controller )
- Centrifuge (Sigma Centrifuge, model: Sigma 1-14 )
Software
- ImageJ imaging software
- SoftWorx software
Procedure
- Preparation of bacteria and media for worm growth
- Bacteria preparation (C. elegans food)
- In 10 ml LB medium, add 10 µl ampicillin (stock 100 mg/ml).
- Inoculate a few colonies of E. coli HT115DE3 empty vector bacteria into the LB + ampicillin medium.
Note: Alternative bacteria: E. coli OP50. - Incubate for 6 h at 37 °C in an orbital shaker at 200 rpm.
- C. elegans growth media preparation
- Melt the solid NGM contained in a glass bottle (500 ml) using a microwave (power level 2 for 25 min).
- Keep the melted medium at 55 °C in a water bath for 15-20 min.
- Before usage, add:
12.5 ml of 1 M KH2PO4 pH 6
500 µl of 1 M MgSO4
500 µl of 1 M CaCl2
500 µl cholesterol (stock 5 mg/ml in 95% ethanol)
500 µl of ampicillin (stock 100 mg/ml), if using knockdown bacteria strain HT115DE3 carrying the L4440 vector or the target gene. - Gently invert the bottle a few times to mix all the ingredients and then pour 6 ml of the medium into each of 60 x 15 mm Petri dish.
- Let it dry at room temperature for around 1 h.
- Once dried, using a sterilized bacterial plating rod spread 70 µl of bacteria prepared in step A1 above to form a lawn. The optical density OD600 of the bacteria is approximately 1.0. Alternatively, the bacteria can be added as 3 drops distributed onto the plate.
- Incubate the plate overnight at 37 °C.
- Melt the solid NGM contained in a glass bottle (500 ml) using a microwave (power level 2 for 25 min).
- Worm growth
- From a grown worm plate, cut 4 pieces of approximately 1 x 1 cm agar with a pointy spatula sterilized with 100% ethanol and flame.
- Transfer each agar piece by turning them over onto each of 4 plates prepared in step A2.
- Incubate these worm plates for 2 days at 20 °C.
- Worm synchronisation
- The 4 plates are expected to be filled with adult worms, visualized through a stereomicroscope, and at which point the synchronization can be done. An alternative method to obtain a huge amount of worms is to grow them in liquid media (NGM medium without agar).
- Add 2 ml of alkaline hypochlorite solution (bleaching solution) to each plate and rapidly collect the worms by pipetting (about 10 sec per plate) in a sterile 15 ml conical tube. Date and label the tube with the name of the worm strain. Alternatively, harvest the worms with 1 ml of M9 buffer and collect them into 15 ml conical tubes. Centrifuge in the Eppendorf 5810 R centrifuge, using swing bucket rotor (A-4-81) with adapters for 15 ml conical tubes at 1,740 x g for 2 min at 4 °C. Discard the M9 buffer and add 2 ml of the bleaching solution. In this way the worms are washed once.
- Quickly start to vortex the worms for 7 min at setting 8 on the vortex and check under the stereomicroscope if the adult worms are lysed and only the eggs are present.
Note: Steps B2 and B3 must be carried out with care as they are critical for the eggs to hatch.
- Without any delay, centrifuge in the Eppendorf 5810 R centrifuge at 1,740 x g for 2 min at 4 °C.
- Aspirate the supernatant, but leave approximately 1 ml of the solution.
- Adding 5 ml of M9 buffer and repeat the centrifugation and aspiration as in steps B4 and B5.
- Repeat 4 more times step B6, in order to remove any trace of the bleaching solution.
- Incubate the eggs in the final 1 ml of M9 buffer overnight at 20 °C. During this time the eggs will hatch and produce L1 stage worms.
- The 4 plates are expected to be filled with adult worms, visualized through a stereomicroscope, and at which point the synchronization can be done. An alternative method to obtain a huge amount of worms is to grow them in liquid media (NGM medium without agar).
- Worm under treatment
- Bacteria preparation
- In 3 different tubes, add 10 ml LB medium and 10 µl ampicillin (100 mg/ml).
- In the 1st tube, add a few colonies of E. coli HT115DE3/empty vector L4440.
- In the 2nd tube, add a few colonies of E. coli HT115DE3/pL4440-oct-1.
- In the 3rd tube, add a few colonies of E. coli HT115DE3/pL4440-oct-2.
- Incubate all the tubes for 5 h in a 37 °C orbital shaker.
- In 3 different tubes, add 10 ml LB medium and 10 µl ampicillin (100 mg/ml).
- Drug plate preparation
- Melt a bottle of 500 ml of solid NGM as above (see C. elegans growth media preparation above).
- For normal drug plates-no knockdown
- Transfer 30 ml of melted NGM media kept at 55 °C into a small sterile bottle. This will make 3 Petri dishes (60 x 15 mm) each containing ~10 ml of NGM media for triplicate assays.
- Add the doxorubicin (stock 2 mg/ml) to the desired final concentrations for each 6 ml of NGM media and pour onto the 60 x 15 mm Petri dish. For doxorubicin use between 0.1 to 100 µM depending on the strains. Uptake was observed with as low as 0.5 µM of doxorubicin.
IMPORTANT: Do not forget to prepare a plate without drug. - Leave plates for at least 15 min at room temperature for the medium to solidify.
- Add 70 µl E. coli HT115DE3/pL4440-empty vector bacteria.
- Incubate the plates overnight at 37 °C.
- Transfer 30 ml of melted NGM media kept at 55 °C into a small sterile bottle. This will make 3 Petri dishes (60 x 15 mm) each containing ~10 ml of NGM media for triplicate assays.
- For knockdown drug plates
- Transfer 30 ml of melted NMG medium kept at 55 °C into a small sterile bottle.
- Add 30 µl of 1 M IPTG, needed to induce the production of dsRNA from the L4440 vector carrying a fragment of the target gene.
- Add to 6 ml of this media the desired concentrations of doxorubicin as above.
IMPORTANT: Do not forget to prepare a plate without drug. - Pour 6 ml of media with and without IPTG and with doxorubicin onto 60 x 15 mm Petri dishes.
- Dry the plates, as above.
- Add 70 µl E. coli HT115DE3/pL4440-oct-1 bacteria for 1st set of 5 plates.
- Add 70 µl E. coli HT115DE3/pL4440-oct-2 bacteria for 2nd set of 5 plates.
- Incubate the plates overnight at 37 °C.
- Melt a bottle of 500 ml of solid NGM as above (see C. elegans growth media preparation above).
- Now all the plates are ready to add 50 µl of the L1 synchronized worms.
- Verify if there are nearly 50 L1-staged worms on the plates under the stereomicroscope.
- Incubate the plates at 20 °C for 3 days.
- Bacteria preparation
- Microscope visualisation
- Preparation of slides
- On a frosted microscope slide add one drop (~50 µl) of 3% agarose and squeeze it with another slide to form a sandwich. After 10 sec, remove one of the slide to expose the agarose.
- Dry the slide with the agarose at 65 °C around 15-30 min using a Programmable Thermal Controller 96 Well.
- On a frosted microscope slide add one drop (~50 µl) of 3% agarose and squeeze it with another slide to form a sandwich. After 10 sec, remove one of the slide to expose the agarose.
- Mounting the worms
- Add 7 µl of 1 M levamisol onto the surface of the dried agar on the slide.
- To each slide, add 10-15 young adult worms from a single treatment condition by using a platinum wire to transfer the worms.
- Sterilize the platinum wire using a flame before transferring the next 10-15 worms from a different treatment condition onto a new slide.
- Add the coverslip and seal around the slide and coverslip with clear nail polish.
- Add 7 µl of 1 M levamisol onto the surface of the dried agar on the slide.
- Microscopy visualisation
The slide containing the worm is ready to be visualised. Place the slide on the stage of an epifluorescence microscope.
1st Adjust the objectives
2nd Select the filters and adjust all the necessary parameters
3rd Once everything is ready, start capturing the images
4th Save the images
Below is a detail description to visualize the worms using a DeltaVision microscope.- Add a small drop of DeltaVision Immersion oil 1.534 onto the slide in order to use objective 40x.
- Place the mounted slide on the microscope by inverting it.
- Adjust the eyepiece filter roll to POL filter.
- Open the SoftWorx software
 . See GE Healthcare manual (GE Healthcare, 2014).
. See GE Healthcare manual (GE Healthcare, 2014). - Press on the microscope.
3 windows will be opened (Figure 1).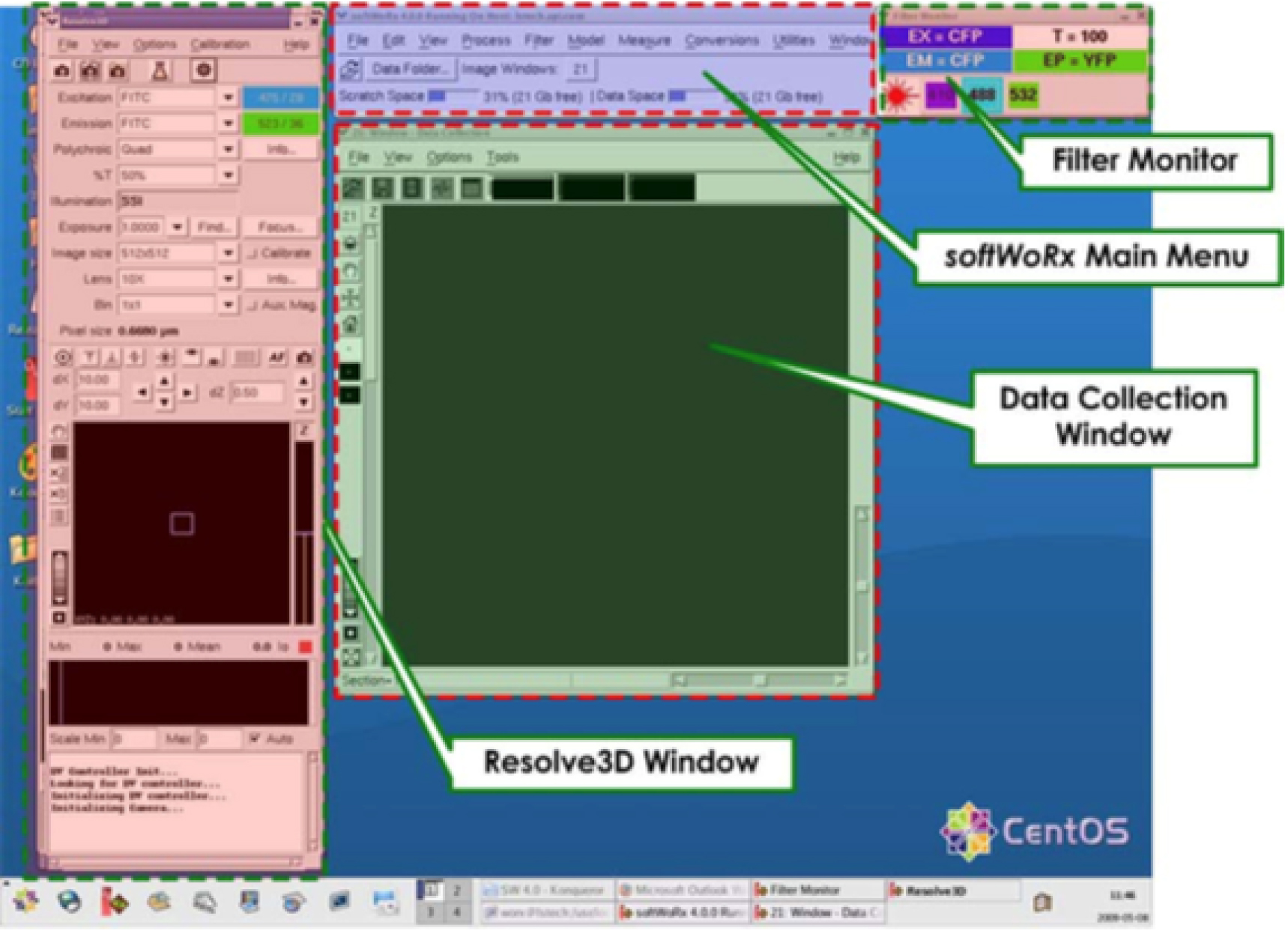
Figure 1. SoftWorx desktop display. The Resolve3D window includes acquisition parameters and controls for moving the stage, the Data Collection window displays images as they are acquired, and the Filter Monitor displays the filters currently selected. - On the window called Resolve3D (Figure 1).
- Press on the gear wheel (setting)
 to open Resolve3D setting window (Figure 2).
to open Resolve3D setting window (Figure 2). 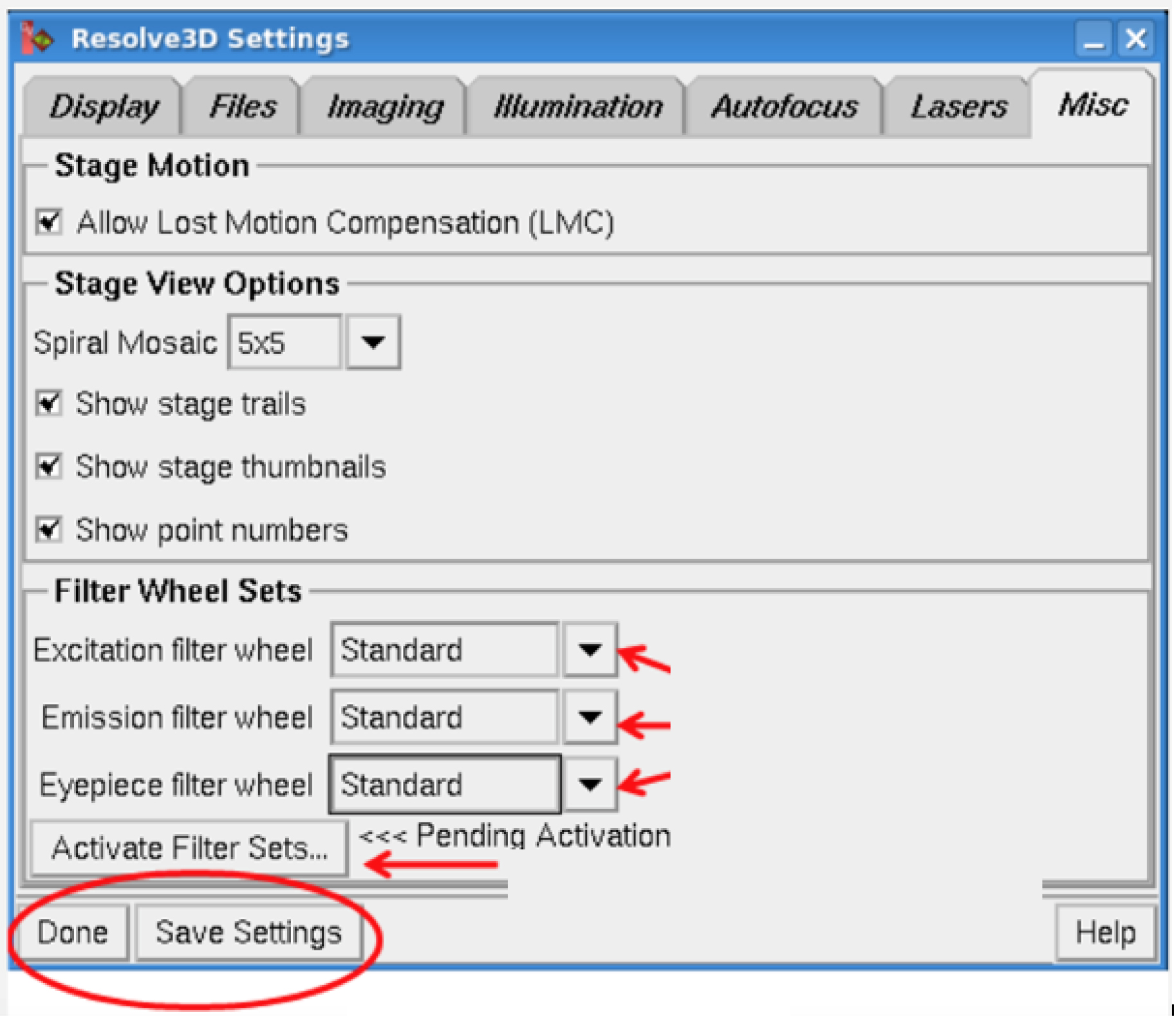
Figure 2. Resolve3D window setting - Under MISC.
- Select the filters.
- Save settings.
- Under FILES (Figure 3).
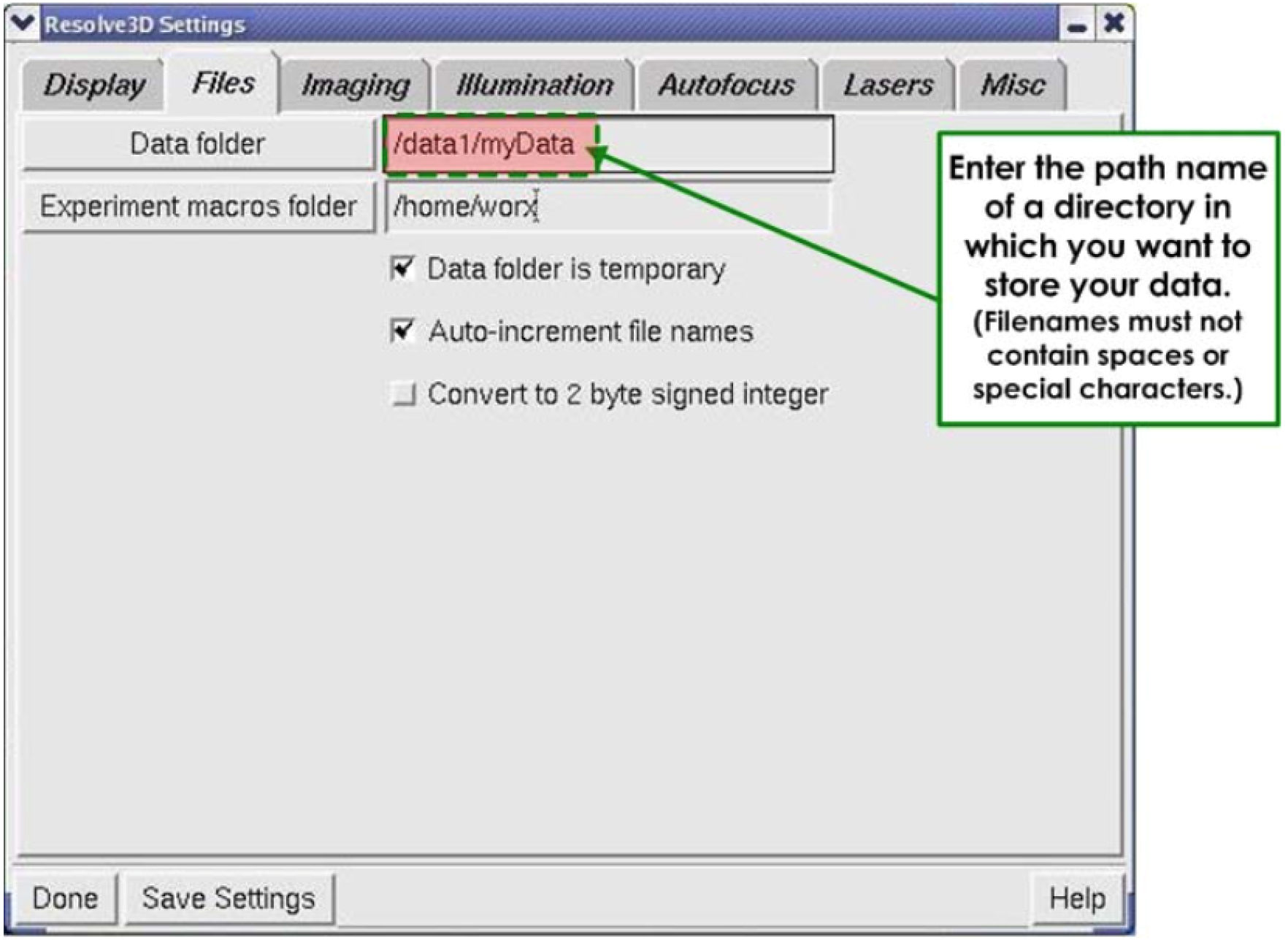
Figure 3. Display of the Files tab - In the Data folder field (Figure 3), enter the directory in which to save the image.
- In the Experiment macros folder field (Figure 3), enter the same directory.
- Save settings.
- Done.
- In the Resolve3D window (Figure 1), set the following parameters in Figure 4.
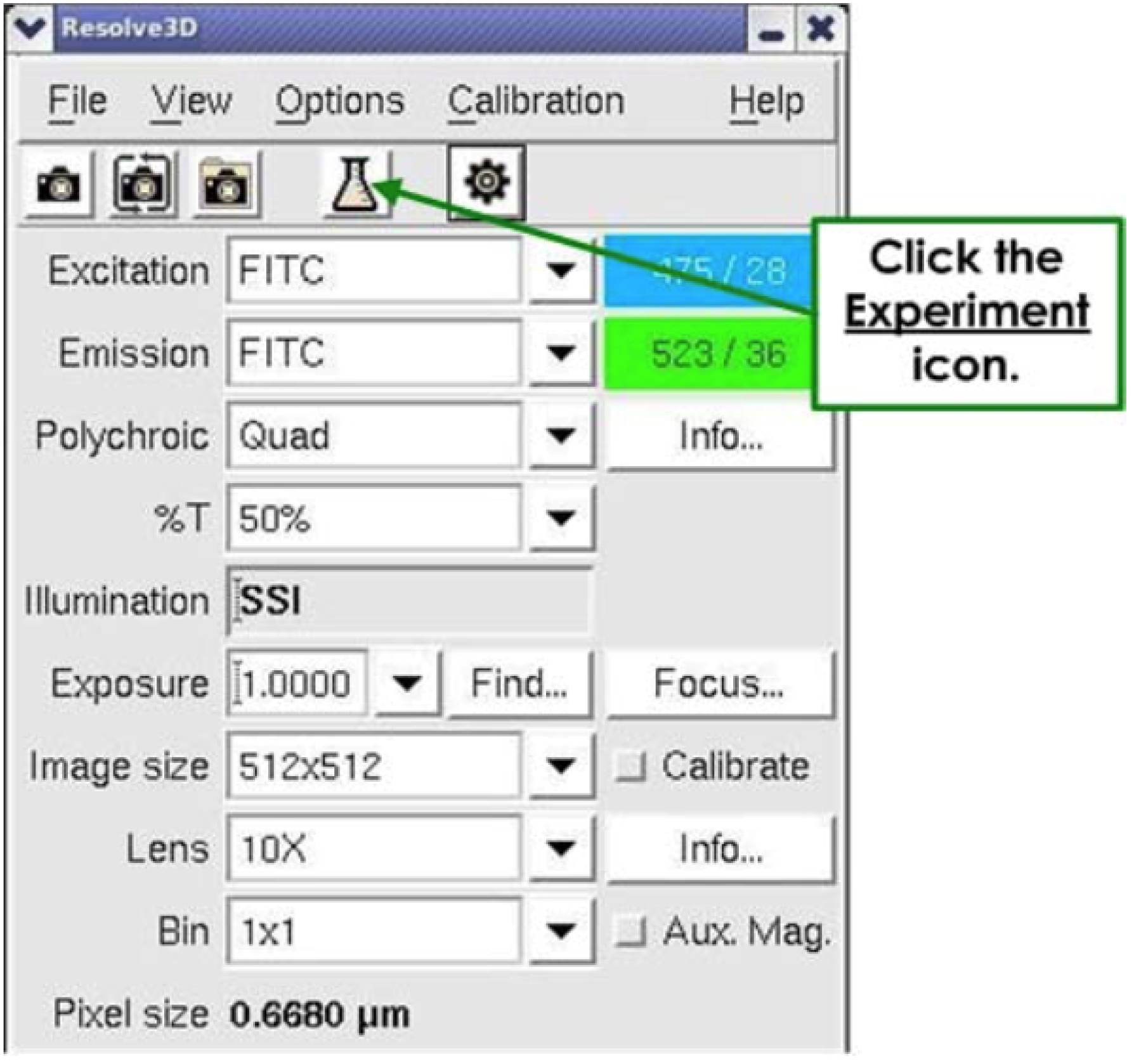
Figure 4. Setting the parameters- Excitation: POL.
- Emission: POL.
- % T: 32%.
- Exposure: 0.025.
- Lens: 40x.
- Excitation: POL.
- Press on the Erlenmeyer.
- A window called Design/Run Experiment window will be opened (Figure 5).
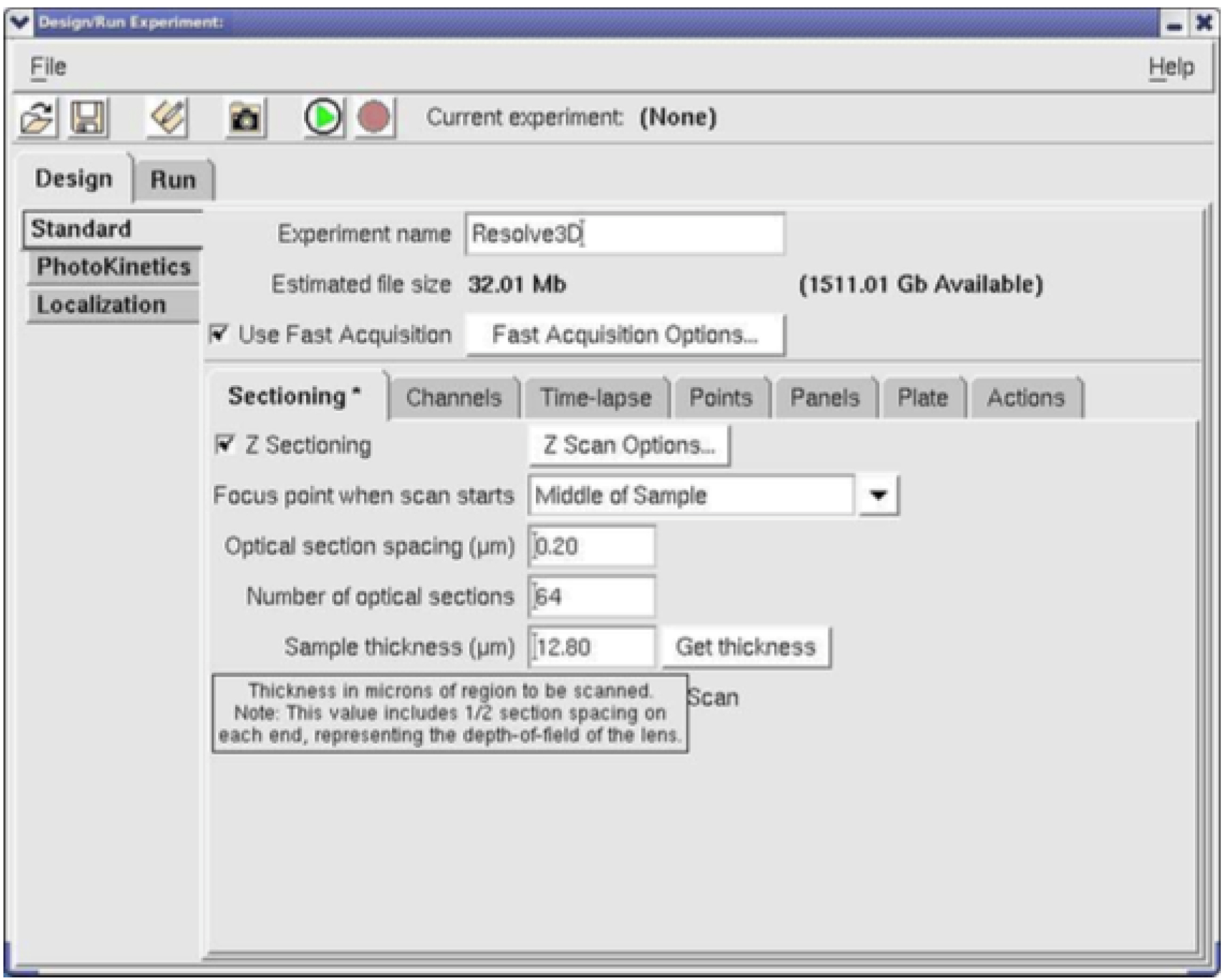
Figure 5. Design and Run experiment window - Under design
1)Experimental name field: write your file name (Figure 5).
2)Under Sectioning
Remove the check beside the Z sectioning.
3)Under channels (Figure 5).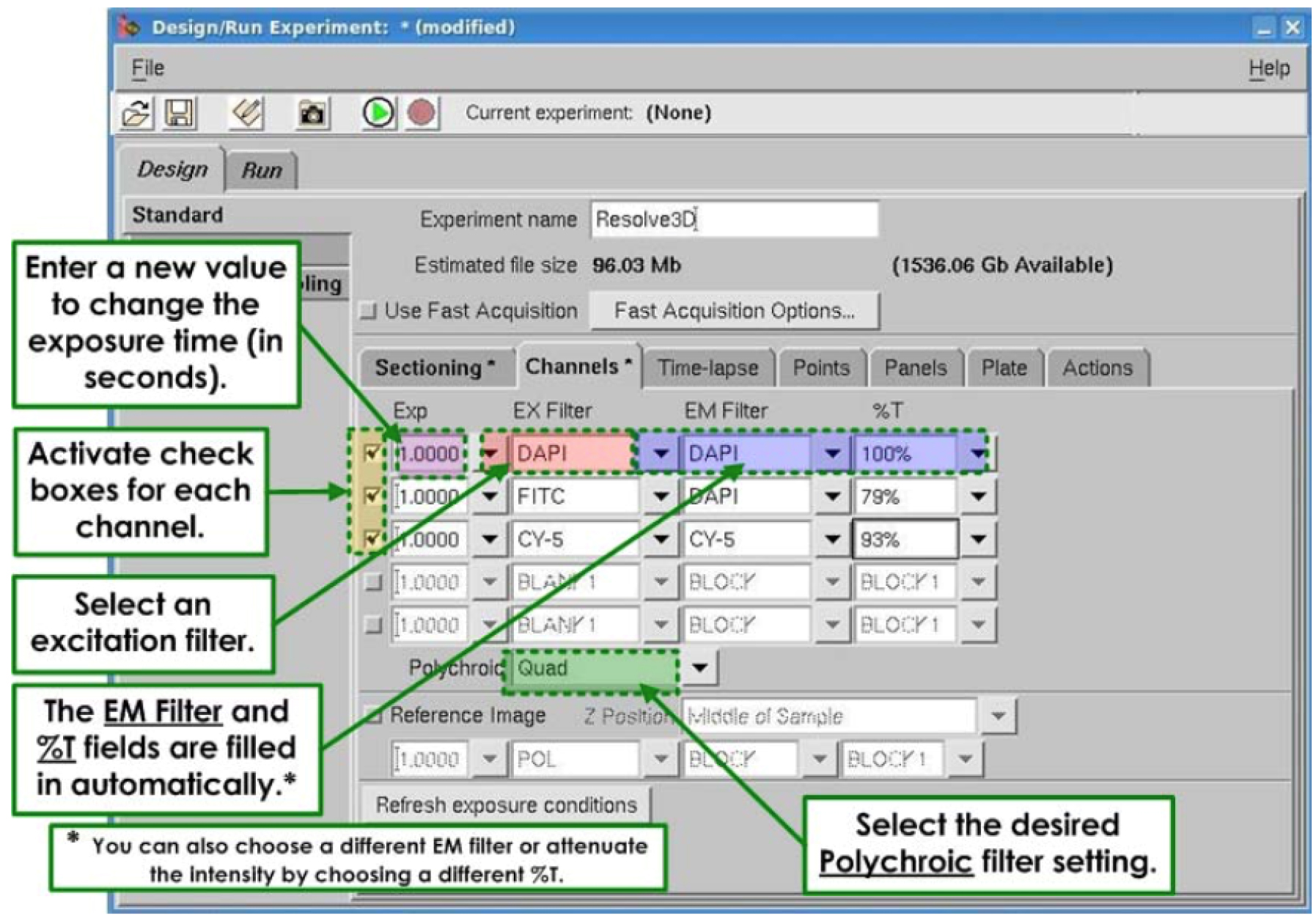
Figure 6. The Channels in the Design and Run experiment windowCheck 1st box (Figure 6)
Under EX filter: POL
Automatically EM filter will be modifying
Under% T: choose 32%
Check 2nd box (Figure 6)
Under EX filter: FITC
Automatically EM filter will be modified
Under% T: choose100%
Check 3rd box (Figure 6)
Under EX filter: mCherry
Automatically EM filter will be modified
Under% T: choose 100% - Under Run before starting to acquire image
Image file name field, write the same name you wrote under the design/Experimental name (Figure 5). - Now you are ready to acquire image by pressing on the start button
 as shown in Figure 7. You can adjust the image by looking on the Resolve3D window which contains the stage trails in the stage View window. This way you can focus at the pharynx level.
as shown in Figure 7. You can adjust the image by looking on the Resolve3D window which contains the stage trails in the stage View window. This way you can focus at the pharynx level.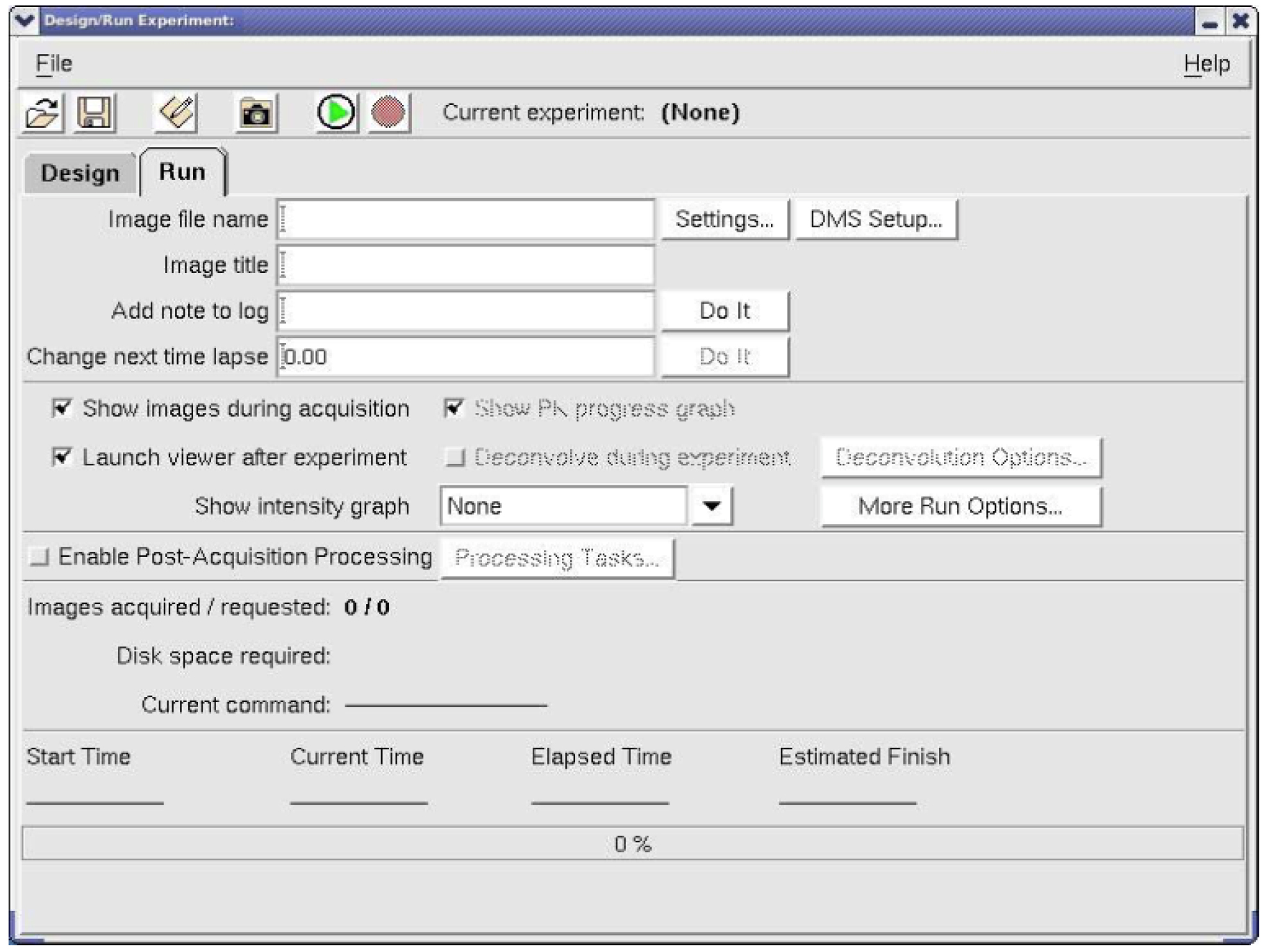
Figure 7. The start button in the Design and Run experiment window
- Add a small drop of DeltaVision Immersion oil 1.534 onto the slide in order to use objective 40x.
- Result analysis
- Software used: ImageJ
- Drag the file to imageJ window.
- Under image → Transform → rotate the image to get an image of the worm placed anterior to posterior.
- Under image → Stacks → Stack to images.
- Under image → Color → merge channels.
Select the image which is appropriate to the desired color. - Create montage: under image → Stacks → make montage.
- Create the 3D picture based on the intensity.
Take the merged image.
Under Plugins → 3D → Interactive 3D surface plot.
- Preparation of slides
Data analysis
The result is representative of a single worm from an experiment where 10 worms were placed onto the slide (Figure 8). The experiment was repeated three times with identical data. Fluorescence posterior to the pharynx is autofluorescence detected from the intestine (Figure 8). The data were quantified using ImageJ according to Papaluca and Ramotar (2016).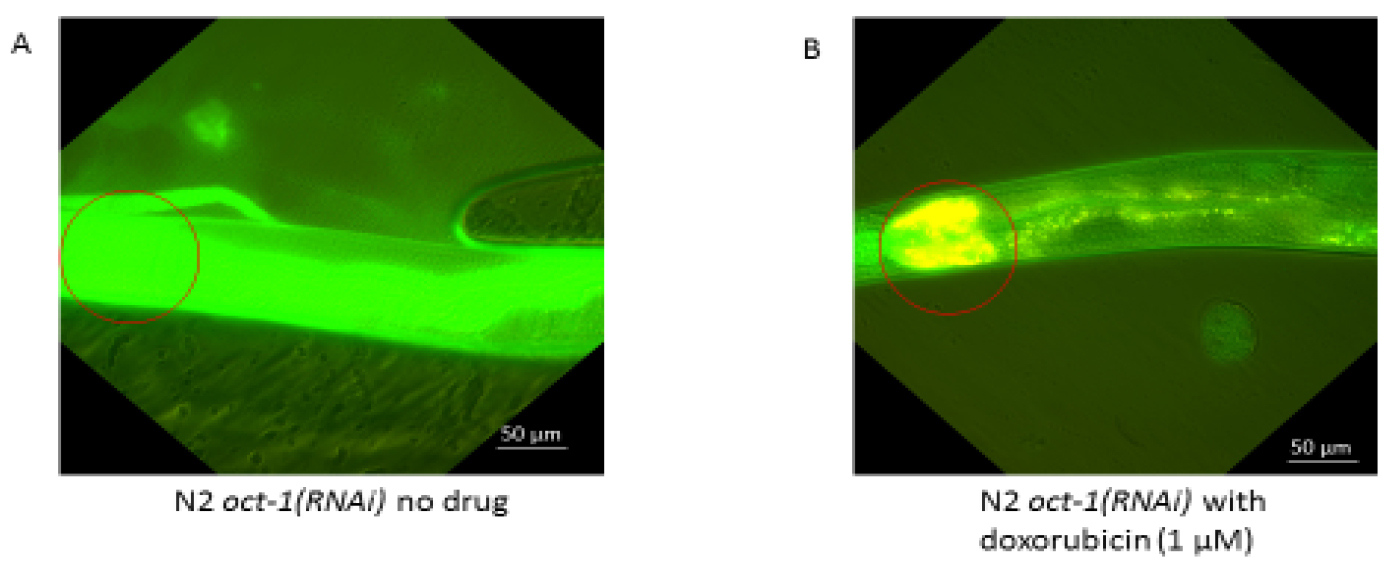
Figure 8. Uptake of doxorubicin in the N2 wild type animals downregulated for oct-1 that causes upregulation of OCT-2. Previously, we showed that downregulation stimulated expression of oct-2, which drives uptake of drugs into C. elegans (Papaluca and Ramotar, 2016). A. Untreated; the absence of doxorubicin is illustrated by the lack of yellow fluorescence and the green background is autofluorescence from ingestion of food by the animal. B. Treated with doxorubicin (1 µM). Doxorubicin uptake is visible as yellow due to the merge of the green autofluorescence and red fluorescence from the drug. Circles show the location of the pharynx. Scale bars = 50 µm. See Data analysis for additional details.
Notes
- Instead of using frozen bacteria as worm food, we typically use fresh culture of the bacteria.
- Always work under sterile conditions. Be on the watch for fungus (white to black like spider net) and rarely mushroom (brownish spots). If this is the case, the best solution will be to bleach the worms and collect the eggs for new larvae.
Recipes
- LB solution
5 g tryptone
2.5 g yeast extract
5 g NaCl
ddH2O up to 500 ml
Add 7.5 g of agar, if making solid media plates - Nematode growth media (NGM)
1.25 g peptone
1.5 g NaCl
8.75 g agar
ddH2O up to 500 ml
Autoclave
When the media is between 50 to 55 °C, add the following constituents before pouring the plates:
12.5 ml of 1 M KH2PO4 pH 6
500 µl of 1 M MgSO4
500 µl of 1 M CaCl2
500 µl cholesterol (stock 5 mg/ml in 95% ethanol)
500 µl of ampicillin (stock 100 mg/ml), if using the plasmid containing bacteria HT115DE3 - Agar pad
3% agarose in MilliQ water
Melt when needed - Alkaline hypochlorite solution (bleaching solution) (make fresh monthly)
8.25 ml ddH2O
3.75 ml 1 N NaOH
3.00 ml Bleach (Javel) - M9 buffer (1 L)
6 g Na2HPO4
3 g KH2PO4
5 g NaCl
0.25 g MgSO4·7H2O
Sterilized by autoclaving
Acknowledgments
This work was funded by the research grant (RGPIN/202432-2012) to D.R. from the Natural Science and Engineering Research Council of Canada. A brief version of this protocol was previously described (Papaluca and Ramotar, 2016).
References
- Burns, A. R., Wallace, I. M., Wildenhain, J., Tyers, M., Giaever, G., Bader, G. D., Nislow, C., Cutler, S. R. and Roy, P. J. (2010). A predictive model for drug bioaccumulation and bioactivity in Caenorhabditis elegans. Nat Chem Biol 6(7): 549-557.
- Cesar-Razquin, A., Snijder, B., Frappier-Brinton, T., Isserlin, R., Gyimesi, G., Bai, X., Reithmeier, R. A., Hepworth, D., Hediger, M. A., Edwards, A. M. and Superti-Furga, G. (2015). A call for systematic research on solute carriers. Cell 162(3): 478-487.
- Cheah, I. K., Ong, R. L., Gruber, J., Yew, T. S., Ng, L. F., Chen, C. B. and Halliwell, B. (2013). Knockout of a putative ergothioneine transporter in Caenorhabditis elegans decreases lifespan and increases susceptibility to oxidative damage. Free Radic Res 47(12): 1036-1045.
- GE Healthcare (2014). DeltaVision Imaging System User’s Manual. PN 29087880 AB.
- O’Reilly, L. P., Luke, C. J., Perlmutter, D. H., Silverman, G. A. and Pak, S. C. (2014). C. elegans in high-throughput drug discovery. Adv Drug Deliv Rev 69-70: 247-253.
- Papaluca, A. and Ramotar, D. (2016). A novel approach using C. elegans DNA damage-induced apoptosis to characterize the dynamics of uptake transporters for therapeutic drug discoveries. Sci Rep 6: 36026.
- Wu, X., Fei, Y. J., Huang, W., Chancy, C., Leibach, F. H. and Ganapathy, V. (1999). Identity of the F52F12.1 gene product in Caenorhabditis elegans as an organic cation transporter. Biochim Biophys Acta 1418(1): 239-244.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Amirthagunabalasingam, S., Papaluca, A., Harihar, T. and Ramotar, D. (2017). Imaging the Pharynx to Measure the Uptake of Doxorubicin in Caenorhabditis elegans. Bio-protocol 7(10): e2291. DOI: 10.21769/BioProtoc.2291.
Category
Cancer Biology > Cancer biochemistry > Genotoxicity
Cell Biology > Cell imaging > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










