- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Comprehensive Methods for Leaf Geometric Morphometric Analyses
Published: Vol 7, Iss 9, May 5, 2017 DOI: 10.21769/BioProtoc.2269 Views: 15856
Reviewed by: Rainer MelzerAnnis Elizabeth RichardsonAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Modified Pseudo-Schiff Propidium Iodide for Staining the Shoot Apical Meristem in Arabidopsis
Ruiqi Li [...] Ligeng Ma
May 5, 2023 2273 Views

A Novel Imaging Protocol for Investigating Arabidopsis thaliana Siliques and Seeds Using X-rays
Brylie A. Ritchie [...] Ansul Lokdarshi
Oct 5, 2023 2189 Views

Direct Plant Regeneration From Immature Male Inflorescence of Banana (Musa spp.)
Pradeep Chand Deo
Oct 20, 2025 1437 Views
Abstract
Leaf morphometrics are used frequently by several disciplines, including taxonomists, systematists, developmental biologists, morphologists, agronomists, and plant breeders to name just a few. Leaf shape is highly variable and can be used for identifying species or genotypes, developmental patterning within and among individuals, assessing plant health, and measuring environmental impacts on plant phenotype. Traditional leaf morphometrics requires hand tools and access to specimens, but modern efforts to digitize botanical collections make digital morphometrics a readily accessible and scientifically rigorous option. Here we provide detailed instructions for performing some of the most informative digital geometric morphometric analyses available: generalized Procrustes analysis, elliptical Fourier analysis, and shape features. This comprehensive procedure for leaf shape analysis is comprised of six main sections: A) scanning of material, B) acquiring landmarks, C) analysis of landmark data, D) isolating leaf outlines, E) analysis of leaf outlines, and F) shape features. This protocol provides a detailed reference for applying landmark and outline analysis to leaf shape as well as describing leaf shape features, thus empowering researchers to perform high throughput phenotyping for diverse applications.
Keywords: Digital leaf morphometricsBackground
There are a variety of approaches to digital leaf shape morphometrics, including outline or Fourier analysis, contour signatures, landmark analysis, shape features, fractal dimensions, and texture analysis (Cope et al., 2012). Among these analyses, landmark and Fourier analysis together perform exceptionally well at distinguishing between groups among leaf shapes (McLellan and Endler, 1998; Hearn, 2009). Landmark analysis is ideal for capturing aspects of shape that are consistent among all leaves within a given dataset. The selection of landmarks should include points that are biologically homologous and adequately represent the morphology of the leaf (see more pointers in Bookstein, 1991 and Zelditch et al., 2004). If leaves in the dataset do not have evolutionarily conserved shape features, ‘pseudo-landmarks’ can instead be placed (Chitwood and Sinha, 2016); that is, landmarks can be placed at equidistant points along the leaf outline relative to homologous points that act as anchors. Landmarks can then be analyzed using Generalized Procrustes Analysis (GPA), which normalizes shape data (annotated by landmarks) at equal scale, allowing for an accurate comparison of shapes regardless of their size. Outline analysis offers a more broadly applicable phenotyping method in that Elliptical Fourier Descriptors (EFDs) are used to build shape descriptors of the leaf outline (Kuhl and Giardina, 1982; Iwata and Ukai, 2002). While sensitive to noise, EFDs are ideal for large leaf datasets that have subtle differences between shapes. Shape features are an additional, simple method of outline analysis that can include the perimeter to area ratio, aspect ratio, and circularity measurements (Cope et al., 2012). In this protocol, we focus on aspect ratio and circularity, as they detect signatures of lobing and serration. Aspect ratio is the ratio of the major axis to the minor axis of a fitted ellipse, in which case values close to ‘1’ are more circular in shape regardless of lobing. Circularity is the ratio of the leaf area to perimeter outline. This measurement is useful for discriminating leaves with lobing and serration, with low circularity values indicating significant lobing and serration. This protocol is designed such that researchers can choose between all three methods (GPA, EFD, and shape features) based on which analyses best fit their data.
Materials and Reagents
- Herbarium specimens and/or fresh leaf material
Equipment
- Computer that can run Microsoft® Windows® XP (or later) and/or Mac® OS X® 10.4 (or later)
- Flatbed photo scanner (Epson Expression, model: 10000XL )
Software
- Adobe® Photoshop® CS4 (or later)
- Epson® Scan Utility v3.4.9.6 (https://support.epson.com/)
- JavaTM (https://java.com/)
- ImageJ (https://imagej.nih.gov/ij/)
- Microsoft® Excel® 2011 (or later)
- SHAPE v1.3 (http://lbm.ab.a.u-tokyo.ac.jp/~iwata/shape/)
SHAPE is built for Windows but if using a Mac, Wine and Winebottler (http://winebottler.kronenberg.org/) are required - R (https://r-project.org/)
Packages: ‘shapes,’ ‘ggplot2,’ ‘devtools,’ ‘ellipse,’ and ‘roxygen2’ - RStudio (https://rstudio.com/products/rstudio/)
RStudio is an optional user interface for R
Procedure
Note: Examples of R scripts and ImageJ macros referenced throughout the protocol can be freely downloaded from GitHub (link: https://github.com/htsvoboda/LeafGeometricMorphometrics.git; Note 1).
- Scanning fresh leaves or herbarium specimens
- For fresh leaves: Place leaves, with petioles removed, flat on the scanner bed. Multiple leaves can be placed on the scanning bed at once, so long as they do not overlap. If the background of the scanner is not already white, place a white piece of paper on top of the leaves; subsequent analyses work best with a solid white background (Note 2).
- For herbarium specimens: carefully place the entire sheet face-down on the scanner bed. Any loose material should be placed in a fragment packet on the sheet.
- Scans should be saved as .jpg or .tif files and named with a unique identifier (e.g., accession number).
Note: It is best to scan at 400 dpi (or a higher resolution) as this improves the quality of the image.
- For fresh leaves: Place leaves, with petioles removed, flat on the scanner bed. Multiple leaves can be placed on the scanning bed at once, so long as they do not overlap. If the background of the scanner is not already white, place a white piece of paper on top of the leaves; subsequent analyses work best with a solid white background (Note 2).
- Acquiring and exporting landmarks
- Create a spreadsheet in Excel with the following column names: ‘order,’ ‘label,’ ‘x,’ and ‘y.’ This file will be referred to as the ‘master spreadsheet.’
- Open ImageJ
For the first use: Select ‘Analyze > Set Measurements…’ and check only the ‘Display label’ checkbox. ‘Redirect’ and decimal place parameters can be left at their defaults. - From the ImageJ menu bar, select the ‘Point selection’ tool (Figure 1; Note 3).
Figure 1. The ImageJ menu bar. Here, the ‘Point selection’ tool button is selected. - Open the first image to be landmarked. In the menu bar, select ‘File > Open...’
- Using the ‘Point selection’ tool in multi-point mode, begin placing landmarks on your predetermined landmark points (see Figure 2; Notes 4 and 5).
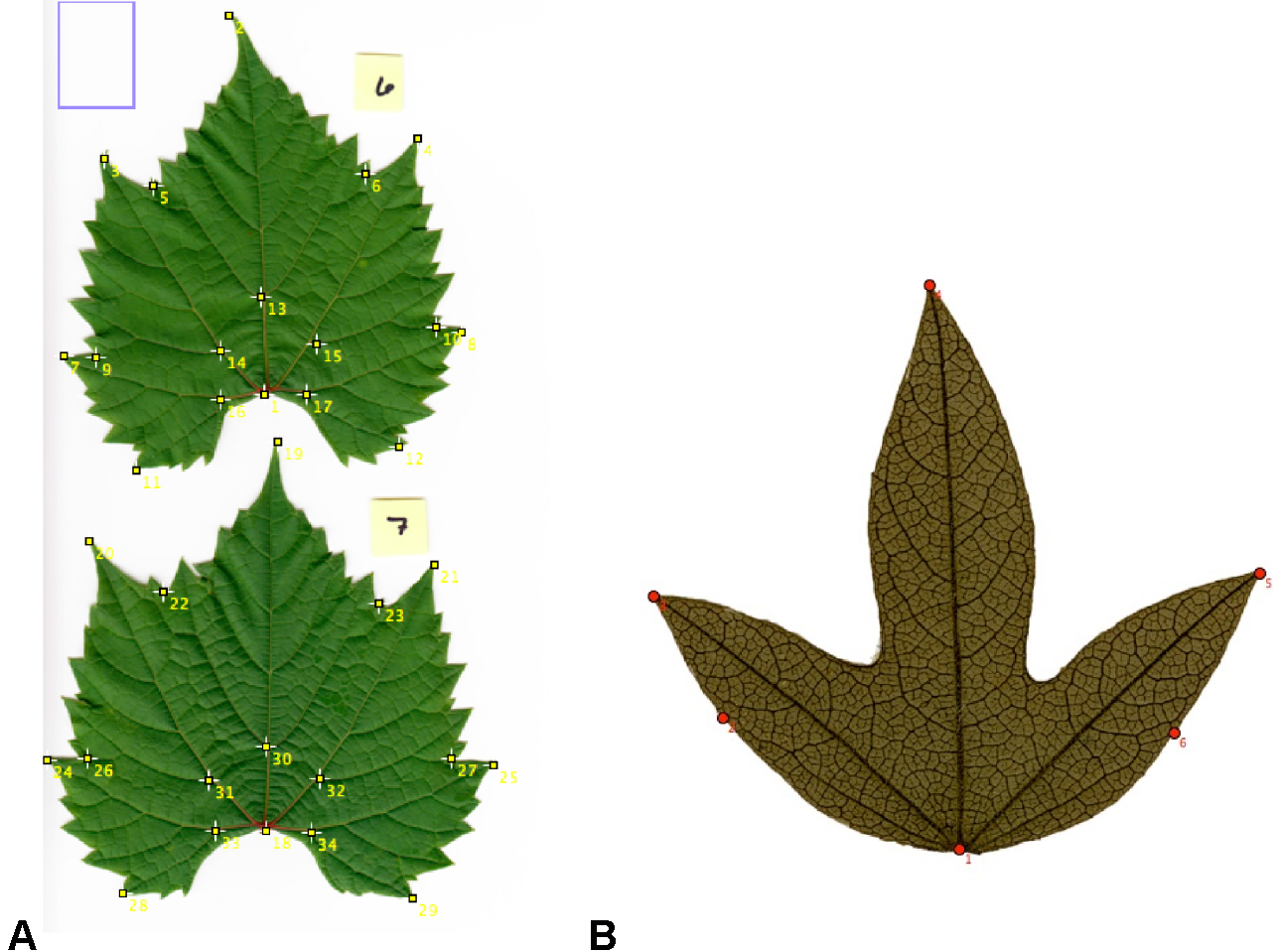
Figure 2. Placement of landmarks on some representative leaves. A. Seventeen landmarks placed on two fresh leaves of Vitis riparia; B. Six landmarks placed on a leaf of Passiflora campanulata from a herbarium specimen. - Once all landmarks have been applied to a leaf (or to multiple leaves per image), view the landmark data. In the menu bar, select ‘Analyze > Measure.’ A new window, called ‘Results’ (Figure 3), will appear with the x, y coordinates for each landmark.
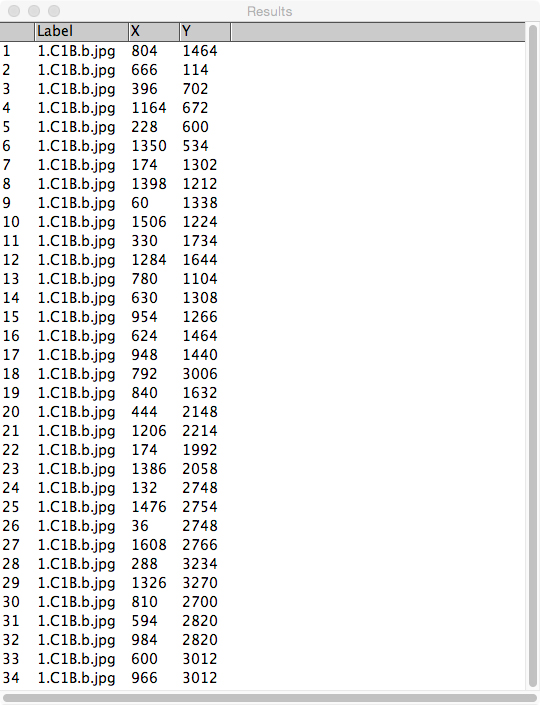
Figure 3. Example of the ‘Results’ window produced by ImageJ. This is viewed by selecting ‘Analyze > Measure’ in the menu bar. - Copy (CTRL + C) and paste (CTRL + V) these data from the ‘Results’ window directly into the ‘master spreadsheet’ (see Figure 4).
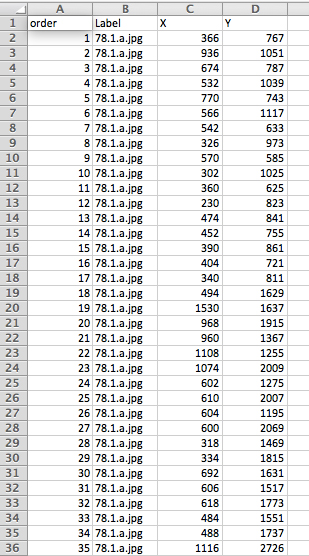
Figure 4. Example format of the ‘master spreadsheet’ - Close the ‘Results’ and image windows and repeat this process with each image file until all leaves have been landmarked. ‘File > Open Next’ can be used if all of the images are in the same folder.
- Landmark coordinates for every leaf will be pasted as-is into the ‘master spreadsheet’. The column ‘order’ may need to be adjusted at the end to reflect a continuous numerical set.
- Before analyzing the landmark data, the ‘master spreadsheet’ will need to be converted from Excel format (.xlsx) to a Tab Delimited Text format (.txt).
- Click ‘File > Save As…’ and select the .txt option.
- Data within the .txt file will need to be reformatted such that the rows are single leaves and the adjacent columns represent the landmark data (x, y coordinates) in sequential order. This can be done in R using the code from GitHub (file: ‘Protocol_stepB-11.R’; R Core Team, 2017).
- Import the reformatted file (referred to as ‘reformatted.txt’) into Excel to verify the data was properly written.
- Click ‘File > Import...’ and choose ‘space’ and/or ‘tab’ as the delimiter (Note 6).
- Supplemental columns and information (i.e., species, genotype, sex, etc.) can be added at this point to help with downstream analyses (Figure 5).
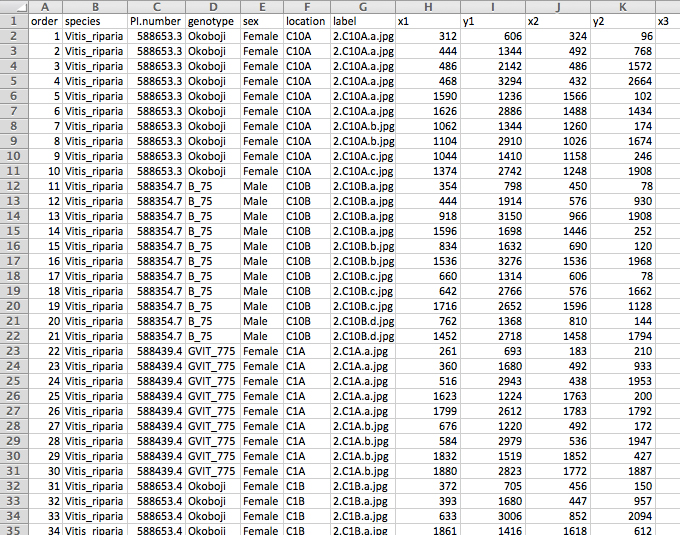
Figure 5. The reformatted ‘master spreadsheet’ (‘reformatted.txt’) as seen in Excel. The spreadsheet now displays each row as a leaf with its landmarks distributed across the columns. Additional information can be added to the reformatted spreadsheet that may be useful in downstream analyses (e.g., columns B-F). - Check landmarks for accuracy. This can be done by plotting the landmark coordinates in R with the package ‘ggplot2’ (Figure 6; Wickham, 2009). The corresponding R script can be downloaded from GitHub (file: ‘Protocol_stepB-14.R’).
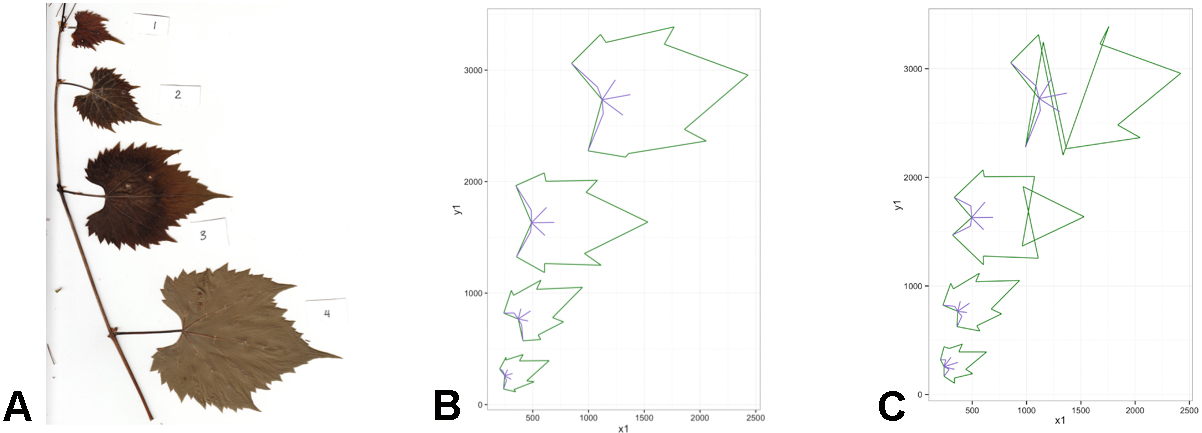
Figure 6. Landmarks are checked for accuracy using the R package ggplot. A. Example of specimen scan; B. The companion plotted landmark coordinates to the scan (A); C. Incorrectly placed landmarks will be immediately apparent visually. Note that (B) and (C) are inverted to that of (A), as pixel coordinates map inversely in a linear regression. - If there appears to be an obvious error in the position of the landmarks for any image (as in Figure 6C), redo landmark placement (step B5) for the affected leaf/file and paste the updated coordinates into the ‘master spreadsheet.’ Repeat steps B11-B12 to reformat and check landmark accuracy for the new spreadsheet.
- Create a spreadsheet in Excel with the following column names: ‘order,’ ‘label,’ ‘x,’ and ‘y.’ This file will be referred to as the ‘master spreadsheet.’
- Analysis of landmarks: Generalized Procrustes Analysis (GPA)
- To perform GPA using the R package ‘shapes,’ (Dryden, 2017) the input file must consist of only landmark data.
- Import (‘File > Import...’) the ‘reformatted.txt’ file into Excel.
- Remove all column headers and any other column information.
- In the menu bar, select ‘File > Save As…’ and rename the file to distinguish that it contains only x, y coordinates (e.g., ‘reformatted_coords.txt’).
- The data can now be processed in R using the ‘shapes’ package.
- The analysis produces Procrustes principal component scores and percent variance explained, Eigenleaves, Eigenvalues, among other informative outputs. The R script includes code that will write files containing PC scores and percentages for further analysis. Example R script can be found on GitHub (file: ‘Protocol_stepC-2-a.R’).
- Because leaf order is preserved in the output files, any additional information (e.g., individual leaf identity, species, genotype, etc.) can be re-added to these output files by adding additional columns to the file, then pasting the additional information for each leaf from the ‘reformatted.txt’ file into the PC score file.
- The leaves can be visualized in morphospace, using the packages ‘ggplot2,’ ‘devtools,’ (Wickham and Chang, 2015) ‘ellipse,’ (Murdoch et al., 2007) and ‘roxygen2’ (Wickham et al., 2015). Example code can be found on GitHub (file: ‘Protocol_stepC-2-c.R’). An example ordination can be seen in Figure 7.
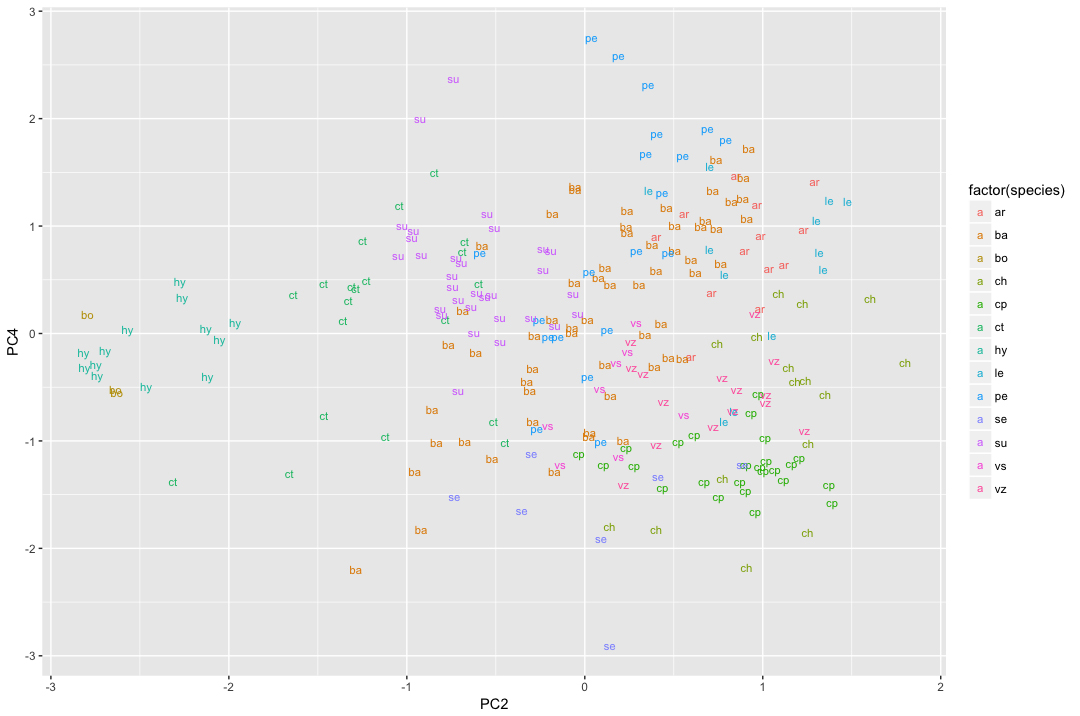
Figure 7. PCA ordination resulting from a Generalized Procrustes Analysis using an example landmark dataset. In this example, the leaves were labeled and color-coded according to the species identity.
- Isolating leaves from scans
- For scans of fresh leaves:
- Open ImageJ.
- Create a macro to isolate leaf images as binary images.
- In the menu bar, select ‘Plugins > New > Macro.’
- A new window will appear titled ‘Macro.txt.’ Input the text from the script found on GitHub (file: ‘Protocol_stepD-1-b-ii.txt’) and leave this window open. We will refer to this macro as the ‘multiple open’ macro (Note 7).
- Next, create a second macro that will select individual leaves from a scan.
- With the ImageJ toolbar as the active window, navigate to the menu bar and select ‘Plugins > New > Macro.’
- A second window will appear titled ‘Macro.txt.’ Input the text from the script found on GitHub (file: ‘Protocol_stepD-1-c-ii.txt’). Leave this macro window open. We will refer to this macro as the ‘select’ macro.
- Open the first image to be analyzed.
- With the ImageJ toolbar as the active window, navigate to the menu bar and select ‘File > Open.’
- Execute the ‘multiple open’ macro by highlighting all code line and clicking either ‘⌘ + R’ (for a Mac) or ‘CTRL + R’ (for a PC) (also see Note 8).
- The leaves within the image should now be thresholded in black and white.
- Isolate single leaves to extract and save as separate files by selecting the ‘Wand (tracing)’ tool from the ImageJ menu toolbar (Figure 8).

Figure 8. The ImageJ menu toolbar. Here, the ‘Wand (tracing)’ tool is selected. - Select an individual leaf by clicking on it and confirm its outline is properly highlighted.
- Highlight all code lines and execute the ‘select’ macro.
- A prompt will appear to save a binary image of a single leaf. Name the file appropriately and save in a different folder with other binary .jpeg files generated during this process for later use.
- Repeat steps D1a-D1h until all leaves in the image have been isolated.
- When each leaf in the open file has been successfully converted to binary images, add the text “run("Open Next");” as the first line of script in the ‘multiple open’ macro.
- Highlight all code lines, including the new line, then execute. The addition of the ‘Open Next’ command will now open the next image file in the folder.
- A prompt will appear to save the changes made to the current open, binary image. Do not save changes, as this will alter the original image scan.
- The next scan will open. Repeat the steps in D1 until all files have been converted to binary images of one leaf per image.
- For scans of herbarium specimens:
- Open Photoshop CS4.
- If the ‘Tools’ menu bar (Figure 9) is not already on the main screen, manually open it by selecting ‘Window > Tools.’
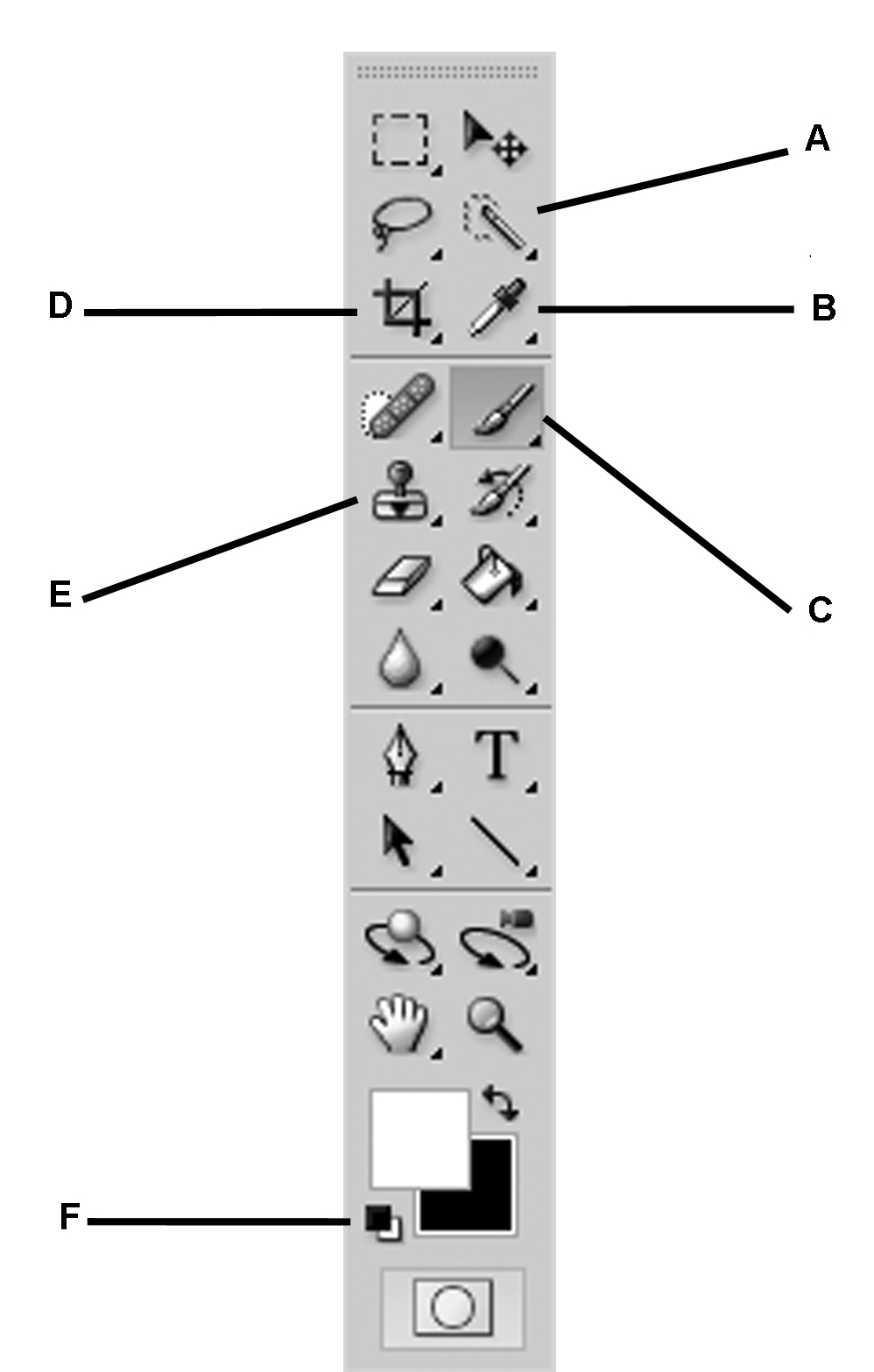
Figure 9. The ‘Tools’ menu in Photoshop. A. Quick Selection Tool; B. Eyedropper Tool; C. Brush Tool; D. Crop Tool; E. Clone Stamp Tool; F. Default Foreground/Background Color Tool. - From the menu, select ‘File > Open…’ and select the first scan to be processed.
- Identify an appropriate leaf (i.e., one that is in good condition, flat, and does not have many [or any] structures intersecting it).
- If the leaf is not in an upright position, rotate it using ‘Image > Image Rotation’ until the apex of the leaf is pointing upward and the base or petiole is pointing downward.
- Use the ‘Crop Tool’ (Figure 9D) to isolate this leaf from most of its surroundings, but leaving a buffer around each side of the leaf.
- If any structures (i.e., stems, tendrils, flowers, other leaves, etc.) intersect the leaf of interest, these must be removed for further analyses.
- Click on the ‘Default Foreground/Background Color Tool’ button (Figure 9F), making sure that ‘white’ is indicated in the foreground box (as seen in Figure 9).
- Select the ‘Brush Tool’ (Figure 9C) and ensure that under the Options Menu (‘Window > Options’) the ‘Mode’ is set to ‘Normal’ and the ‘Opacity’ is set to ‘100%’.
- Using the ‘Brush Tool’, select an appropriate brush diameter and then paint over any structures or tissues that intersect with the leaf of interest (Figure 10).

Figure 10. An example of areas edited out using the ‘Brush Tool.’ By using this tool in Photoshop, intersecting structures have been painted over to remove them.
- Click on the ‘Default Foreground/Background Color Tool’ button (Figure 9F), making sure that ‘white’ is indicated in the foreground box (as seen in Figure 9).
- Click on the ‘Quick Selection Tool’ (Figure 9A). Left-click on the leaf and drag along the blade until the entire lamina, and only the lamina, is outlined.
- Right-click in the selected area and then click ‘Select Inverse’ from the new menu.
- Switch back to the ‘Brush Tool’ and increase the brush diameter. Paint over the background (with white) until only the leaf remains.
- Right-click the image and select ‘Select Inverse’ again.
- Any holes, tears, or anomalies on the leaf surface need to be filled in so as to match the color of the lamina. This can be done using either the ‘Clone Stamp Tool’ (Figure 9E) or the ‘Eyedropper Tool’ (Figure 9B).
- To use the ‘Clone Stamp Tool,’ hold the Alt/Option key and click an intact area on the leaf. Now use the tool to fill in any damaged spots.
- To use the ‘Eyedropper Tool,’ left-click on an intact area of the leaf near the damaged area to extract the color. Switch to the ‘Brush Tool’ and paint over the damaged area(s) to match the color of the rest of the leaf.
- In some cases, especially when using .tif files, it may be necessary to ‘flatten’ the layers of the image by clicking ‘Layers > Flatten Image’ before saving. This will merge all of the edits into a single, savable image.
- Save each leaf (‘File > Save as…’) with a slightly different filename, but still keeping track of the original scan that it came from (e.g., ‘MO1624745_leaf1,’ ‘MO1624745_leaf2,’ etc.).
- Repeat steps D2a-D2n for multiple leaves per scan, and for multiple scans.
- To convert isolated leaf outlines to black and white images for SHAPE analysis (step D3), follow steps D1a-D1f.
- The program SHAPE (Iwata and Ukai, 2002) uses binary leaf outline image files in BMP format. Leaf images can be converted in ImageJ by creating a batch macro.
- Open ImageJ.
- Create a macro to easily convert many images at once.
- From the ImageJ menu click ‘Process > Batch > Macro...’
- Select the appropriate ‘Input’ folder containing the images to be converted.
- Select an ‘Output’ folder to contain the new binary images.
- Choose ‘BMP’ from the ‘Output Format’ drop-down menu.
- Input the text from the script found on GitHub (file: ‘Protocol_stepD-3-b-v.txt’) into the large blank space provided.
- From the ImageJ menu click ‘Process > Batch > Macro...’
- View the output folder to check that the .bmp files were properly converted (Note 9).
- Open ImageJ.
- Analysis of outlines: Elliptical Fourier Descriptors (EFDs)
- Use the SHAPE software to convert image outlines to chain code. We encourage users to become familiar with the SHAPE User Manual in order to better explore parameter choice.
- Open the ChainCoder program within SHAPE.
- Select ‘Files > Select Image File(s).’
- In the following window (Figure 11), select the folder of BMP files. The BMP files will then appear in the field ‘File(s).’
- Select all images and select the double arrow button to transfer the desired files into the ‘Selected File(s)’ field. Press ‘OK.’
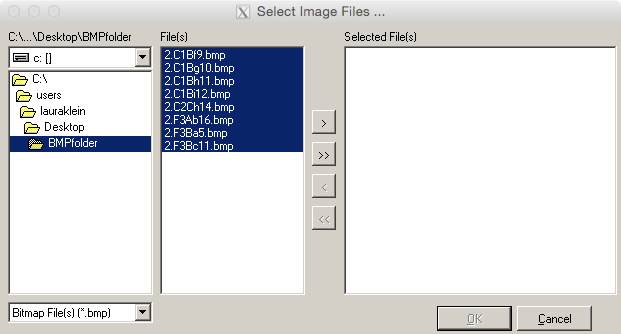
Figure 11. Selection of .bmp files for chain coding in the software SHAPE. Use the double arrow button to move all files to the ‘Selected File(s)’ field.
- Select ‘Files > Select Image File(s).’
- Before beginning the analyses, select the ‘Config’ tab (Figure 12).
- Set ‘Object Color’ to ‘Dark (Black),’ and ‘Scale Included’ to ‘No.’ Leave the other fields at their defaults.
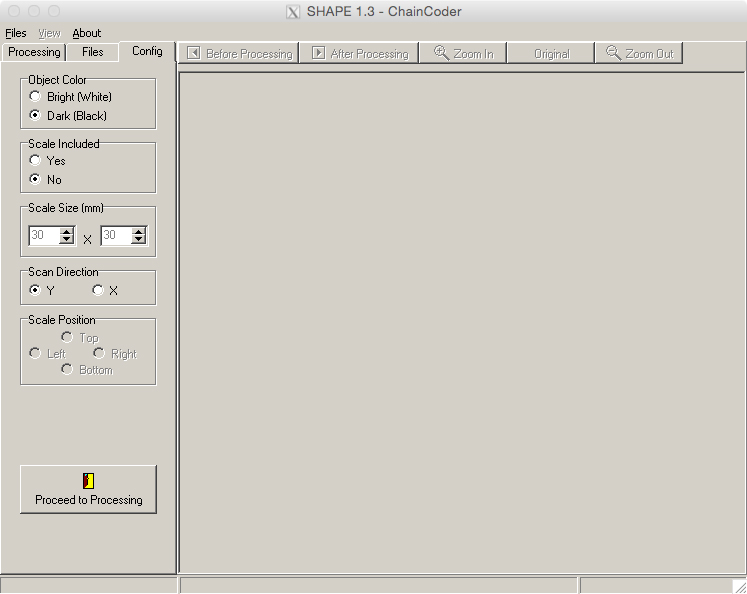
Figure 12. The ‘Config’ tab of the ChainCoder program. This allows for configuration of parameters before beginning the chain coding process.
- Set ‘Object Color’ to ‘Dark (Black),’ and ‘Scale Included’ to ‘No.’ Leave the other fields at their defaults.
- Select the ‘Processing’ tab to begin processing photos.
- Select ‘Load Image’ (Note 10).
- Deselect the ‘Select Area’ box.
- Select ‘Gray Scale.’
- Select ‘Make Histogram.’
- Select ‘Binarize Image.’
- Check the ‘Ero Dil Filter’ and ‘Dil Ero Filter’ boxes and select to what degree to ‘erode’ and ‘dilate’ the outline; this option adds and subtracts the amount of pixels from the image to give a smoother border.
- Select the ‘Labeling Object’ button. A new window named ‘Chain Code Data’ will appear, allowing the user to indicate which objects, over a certain number of pixels, should be isolated for analysis.
- Select ‘Chain Coding.’ This will add the chain code to the user selection.
- Select ‘Save to File.’ For the first image, you will have to name the file. Subsequently, chain codes for further images will be saved as processed (Note 11).
- Select ‘Load Image’ (Note 10).
- Repeat steps E1a-E1c for all further images. If many images need to be processed, hold down the ‘enter’ key (or put a weight on it until the image processing is finished).
- Open the ChainCoder program within SHAPE.
- Convert chain code to normalized EFDs.
- Open the CHC2NEF program within SHAPE. This program converts the chain code created above to normalized EFDs.
- A new window will appear (Figure 13). Select the chain code file produced in step E1c.ix (‘CHC File Name’), then create a name for the new NEF file that will be generated in the following steps (‘NEF File Name’).
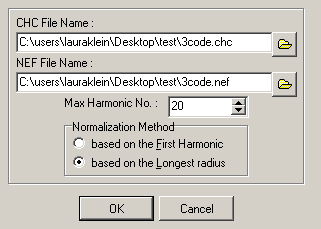
Figure 13. The CHC2NEF window. Select the chain code input file (.chc) and name the resulting normalized elliptical Fourier descriptors file (.nef). - Set the ‘Max Harmonic No.’ Higher harmonic numbers lead to better shape approximations, but usually 20 is sufficient to recapitulate leaf shape accurately.
- Select the Normalization Method to be ‘based on the longest radius.’ This is the way the image will initially be oriented, and this option allows better subsequent manipulation to align properly.
- Click ‘OK.’
- A new window will appear. Click ‘Start !!’
- Orient the image so that all images are similarly aligned. This is an arbitrary designation, but needs to be consistent among all images.
- There are a number of buttons to assist in this process (see Figure 14). The image can be turned by any number of degrees in either direction, and arrow buttons can be adjusted by rotation degree. Alternatively, images can rotate by 180°.
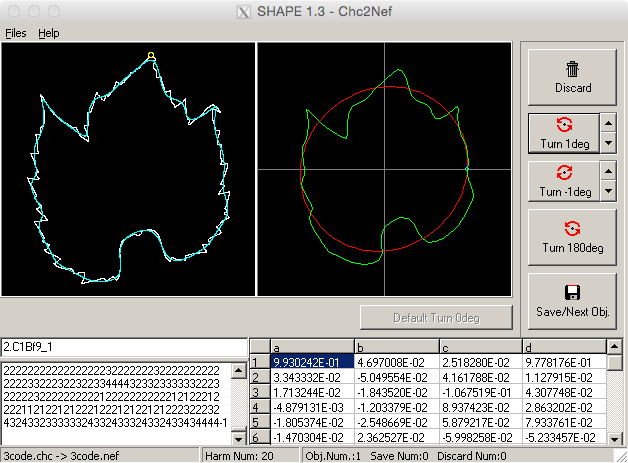
Figure 14. Orienting leaf image chain code to normalized elliptical Fourier descriptors. Utilize the three turning buttons at the right side of the screen to orient the leaf image appropriately. NEF code will reflect these changes (bottom of window). - Once aligned, click ‘Save/Next Obj.’ and repeat until all images have been normalized.
- There are a number of buttons to assist in this process (see Figure 14). The image can be turned by any number of degrees in either direction, and arrow buttons can be adjusted by rotation degree. Alternatively, images can rotate by 180°.
- Open the CHC2NEF program within SHAPE. This program converts the chain code created above to normalized EFDs.
- Visualize Elliptical Fourier Descriptors (EFDs) using Principal Component Analysis (PCA). These analyses can be done within the SHAPE program itself (see below; Note 12).
- Open the ‘PrinComp’ program within SHAPE.
- A new window will appear. In the menu bar, select ‘Files > Open Nef File’ and select the desired .nef file.
- An additional window will appear (Figure 15) with parameters to determine for the PCA.
Figure 15. The ‘NEF File Information’ window. Desired PCA parameters can be set for the normalized Elliptical Fourier Descriptors.- Number of Header Lines’ is automatically set according to the NEF file.
- Select an appropriate number of harmonics on which to perform the PCA (the default is 20).
- Select the desired coefficients to keep constant.
1)To analyze both symmetric and asymmetric variance: select ‘a-d.’
2)To analyze symmetric variance: select ‘a’ and ‘d.’
3)To analyze asymmetric variance: select ‘b’ and ‘c.’
- Number of Header Lines’ is automatically set according to the NEF file.
- Perform the PCA by clicking the ‘Principal Component Analysis’ button (see Figure 16).
Figure 16. The PrinComp program toolbar. A. ‘Principal Component Analysis’ button performs the PCA; B. ‘Calculate Principal Component Scores’ button generates PC scores for the dataset; C. ‘Reconstruct Principle Component Contours’ button generates Eigenleaves for visualization. - Verify that parameters are correct in a new window, and click ‘OK.’
- Once the PCA results have been generated, a new window will appear to save the results file (.pcr file) in the desired folder.
- A new window will appear with information from the PCA (Figure 17). There are a number of tabs with useful information about the analysis.
Figure 17. The ‘Information of Principal Component Analysis’ window. This window will appear after the program has completed the PCA run, providing useful information about the results of the analysis in various tabs.- At the bottom of the window, select the ‘Make Report’ button.
- A new window titled ‘Report Option Dialog’ (Figure 18) will appear to select analysis information that will be written into a report (.txt file). Confirm the ‘Eigenvalues & Eigenvalue Proportions’ box is checked, as this contains the percent variance explained by each principal component (PC).
Figure 18. Select report information for printing. Boxes checked in this window will include relevant information in a report that will be written as a .txt file. - Click ‘OK.’ A new window will appear with the text file containing a PCA report.
- At the bottom of the window, select the ‘Make Report’ button.
- To retrieve the PC values for further analysis, click the ‘Calculate Principal Component Scores’ button in the PrinComp toolbar (Figure 16B) to create a PC score file.
- A new window will appear to name and select the output file. Click ‘OK.’
- To visualize the ‘Eigenleaves’ and what each PC represents, click the ‘Reconstruct Principal Component Contours’ button in the PrinComp toolbar (Figure 16C).
- A dialogue box will appear to select the number of components to reconstruct, options being ‘Reconstruct Effective Components Only’ or ‘Select Manually...’ (Figure 19). This will be the number of components that will be visualized.
Figure 19. The ‘Reconstruct Contours’ window. In order to visualize the Eigenleaves, select the desired number of components to be reconstructed.
- A dialogue box will appear to select the number of components to reconstruct, options being ‘Reconstruct Effective Components Only’ or ‘Select Manually...’ (Figure 19). This will be the number of components that will be visualized.
- Save the resulting ‘PC contours’ file. New windows will appear (opens automatically in SHAPE’s PrinPrint program; Figure 20), one with a graphic showing Eigenleaves, another to select draw options.
- The PrinPrint program can be used to view the ‘PC contours’ file at a later time.
Figure 20. Eigenleaves visualized in the ‘PrinPrint’ program
- Use the SHAPE software to convert image outlines to chain code. We encourage users to become familiar with the SHAPE User Manual in order to better explore parameter choice.
- Shape features: Aspect ratio and circularity
- Open ImageJ.
- Navigate to the menu bar and select ‘Analyze > Set Measurements...’
- Check the ‘Area,’ ‘Shape Descriptors,’ and ‘Display label’ boxes.
- Navigate to the menu bar and select ‘Process > Batch > Macro...’
- A new window will appear titled ‘Batch Process.’ Select the appropriate input (i.e., binary leaf image files) and output folders (Note 13).
- In the macro field, input the ImageJ code found on GitHub (file: ‘Protocol_stepF-4-b.txt’).
- Select ‘Process.’
- A new window will appear titled ‘Results.’ Save this report.
- The resulting data can be visualized using linear regression (Figure 21).
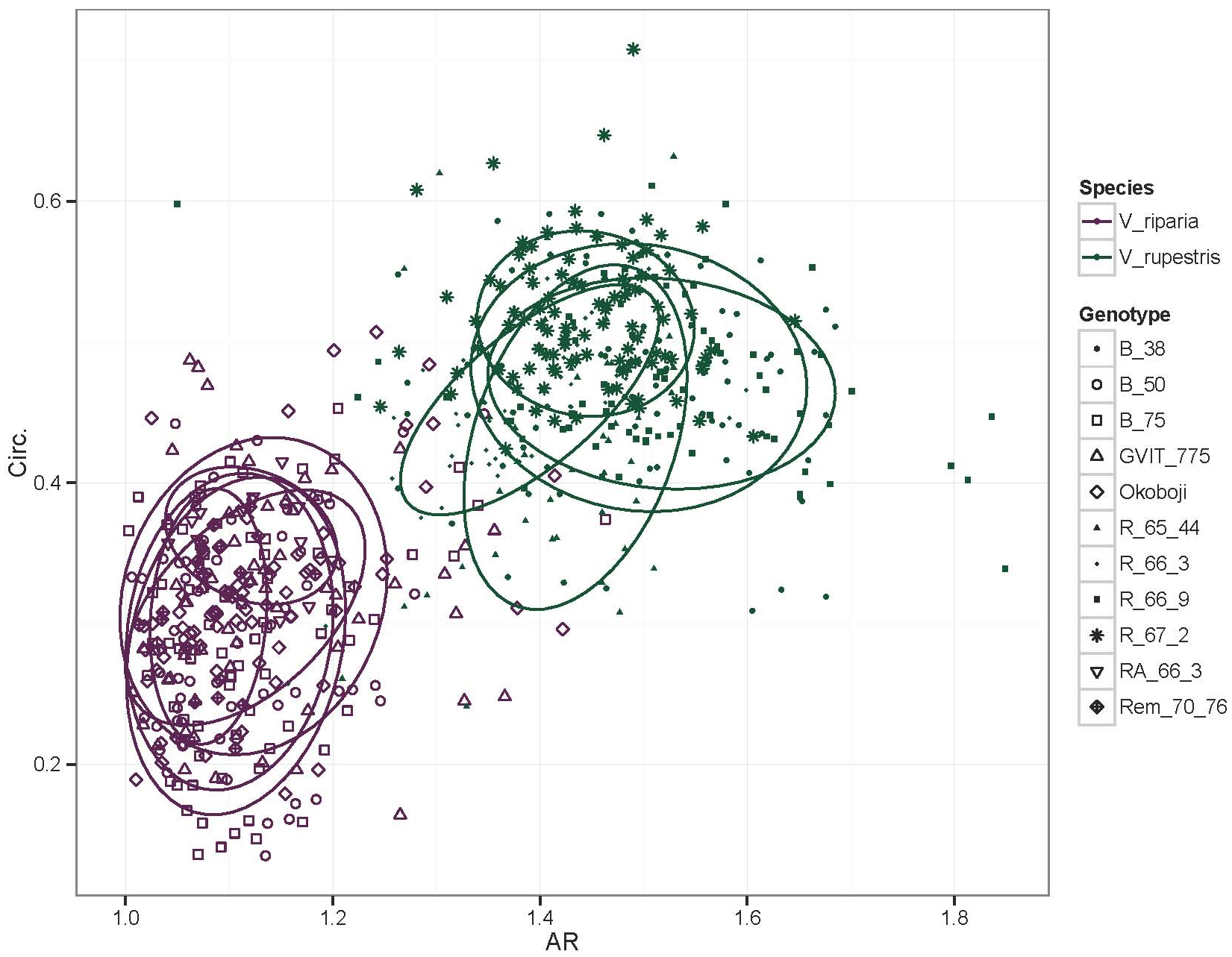
Figure 21. Linear regression of Aspect Ratio (AR) and Circularity (Circ.) data. In this example, multiple leaves from several genotypes of Vitis riparia (purple) and V. rupestris (green) have been measured for AR and Circ., with the resulting data visualized in this linear regression. Low AR and Circ. values capture the deeper lobing and significant serration of V. riparia leaves compared to that of V. rupestris leaves.
- Open ImageJ.
Data analysis
Data analysis techniques are fundamental to the purpose of this protocol and are integrated within the procedure (i.e., C, E3, F5), as they are often challenging for new users to develop de novo. However, our methods represent a sampling of available methods for digital morphometric analysis. We encourage users to explore the literature and available programs to develop a method that is best suited for their material and the particular scientific inquiry.
Notes
- In the scripts available from GitHub, any text preceded with ‘#’ should be considered user notes–these will not be read by the computer if the whole script is copied and pasted into the R or macro consoles.
- Because Generalized Procrustes Analysis (GPA) and outline analysis do not require scaling, it is not necessary to include a ruler in scans for morphometric analyses; however, we recommend this as good practice.
- If the point tool is in the single ‘Point’ setting, simply right-click on the button to change it to the ‘Multi-point’ setting.
- Landmarks must be placed in the same order on all leaves.
- If an error was made during placement, points can be deleted by holding the CTRL key and clicking on the point. It is also possible to move points by clicking on the imprecise point, then moving it to the desired location.
- It may be necessary to adjust column headers or other information that might be erroneously askew from the conversion process.
- If there is only one fresh leaf per scan, refer to the directions in step D1e-D1f. Repeat until all images have been converted to black and white.
- For the first image file of the dataset, do not use the first line of code (e.g., run(“Open Next”)).
- BMPs are large files, and it may be convenient to use an external hard drive or cloud storage client to store and use them from this point forward.
- Viewing the first image in the viewing field may require using the ‘Zoom Out’ button in the top right corner to visualize properly.
- This produces one file for all of the images’ chain code. If chain-coding cannot be completed in one sitting, the file can be updated to include the remaining files at a later date.
- Alternatively, using the R package ‘Momocs’ (Bonhomme et al., 2014) converts NEFs to objects for a variety of graphical visualizations, including PCA.
- The desired output will not be a folder of files, rather the measurement report that can be saved as a single file. Therefore, the output folder can be the same as the input folder. To minimize file size, select a small file format (we suggest .jpeg) in the ‘Output Format’ drop down menu.
Acknowledgments
This protocol was developed in part for the publications of Chitwood et al. (2016) and Klein et al. (2017). The authors are grateful to Dr. Dan Chitwood for his comments, guidance, and expertise in using and developing these methods. We would also like to thank Dr. Allison Miller lab undergraduate members for their comments and suggestions on the protocol, as well as three reviewers who helped improve this manuscript. A Saint Louis University Graduate Research Assistantship to LLK and an Ohio University Original Work Grant to HTS supported this work.
References
- Bonhomme, V., Picq, S., Gaucherel, C. and Claude, J. (2014). Momocs: Outline analysis using R. J Stat Softw 56(1): 1-24.
- Bookstein, F. L. (1991). Morphometric Tools for Landmark Data: Geometry and Biology. Cambridge University Press, pp: 435.
- Chitwood, D. H., Klein, L. L., O’Hanlon, R., Chacko, S., Greg, M., Kitchen, C., Miller, A. J. and Londo, J. P. (2016). Latent developmental and evolutionary shapes embedded within the grapevine leaf. New Phytol 210(1): 343-355.
- Chitwood, D. H. and Sinha, N. R. (2016). Evolutionary and environmental forces sculpting leaf development. Curr Biol 26(7): R297-306.
- Cope, J. S., Corney, D., Clark, J. Y., Remagnino, P. and Wilkin, P. (2012). Plant species identification using digital morphometrics: A review. Expert Syst Appl 39(8): 7562–7573.
- Dryden, I. L. (2017). Shapes: Statistical Shape Analysis. R package version 1.1-8.
- Hearn, D. J. (2009). Shape analysis for the automated identification of plants from images of leaves. Taxon 58(3): 934-954.
- Iwata, H. and Ukai, Y. (2002). SHAPE: a computer program package for quantitative evaluation of biological shapes based on elliptic Fourier descriptors. J Heredity 93: 384-385.
- Klein, L. L., Caito, M., Chapnick, C., Kitchen, C., O’Hanlon Regan, Chitwood, D. H. and Miller, A. J. (2017). Digital morphometrics of two North American grapevines (Vitis: Vitaceae) quantifies leaf variation between species, within species, and among individuals. Front Plant Sci 8: 373.
- Kuhl, F. P. and Giardina, C. R. (1982). Elliptic features of a closed contour. Comput Vision Graphs 18: 236-258.
- McLellan, T. and Endler, J. A. (1998). The relative success of some methods for measuring and describing the shape of complex objects. Sys Biol 47(2): 264-281.
- Murdoch, D., Chow, E. D. and Celayeta, J. F. (2007). ellipse: Functions for drawing ellipses and ellipse-like confidence regions. R package version 0.3-5.
- R Core Team. (2017). R: a language and environment for statistical computing. R foundation for statistical computing. Vienna.
- Wickham, H. (2009). ggplot2: Elegant Graphics for Data Analysis. Springer.
- Wickham, H. and Chang, W. (2015). devtools: Tools to Make Developing R Packages Easier. R package version 1(0): 185.
- Wickham, H., Danenberg, P. and Eugster, M. (2015). roxygen2: In-source documentation for R, 2015. R package version 5(1).
- Zelditch, M. L., Swiderski, D. L., Sheets, H. D. and Fink, W. L. (2004). Geometric Morphometrics for Biologists: A Primer. Elsevier Academic Press, pp: 437.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Klein, L. L. and Svoboda, H. T. (2017). Comprehensive Methods for Leaf Geometric Morphometric Analyses. Bio-protocol 7(9): e2269. DOI: 10.21769/BioProtoc.2269.
- Chitwood, D. H., Rundell, S. M., Li, D. Y., Woodford, Q. L., Yu, T. T., Lopez, J. R., Greenblatt, D., Kang, J. and Londo, J. P. (2016). Climate and Developmental Plasticity: Interannual Variability in Grapevine Leaf Morphology. Plant Physiol 170(3): 1480-1491.
Category
Plant Science > Plant developmental biology > Morphogenesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









