- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Metabolic Heavy Isotope Labeling to Study Glycerophospholipid Homeostasis of Cultured Cells
Published: Vol 7, Iss 9, May 5, 2017 DOI: 10.21769/BioProtoc.2268 Views: 7439
Reviewed by: Neelanjan BoseMichael EnosAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Lipidomics Workflow for Analyzing Lipid Profiles Using Multiple Reaction Monitoring (MRM) in Liver Homogenate of Mice with Non-alcoholic Steatohepatitis (NASH)
Hai Ning Wee [...] Jianhong Ching
Jul 5, 2023 2459 Views

Computational Analysis of Plasma Lipidomics from Mice Fed Standard Chow and Ketogenic Diet
Amy L. Seufert [...] Brooke A. Napier
Sep 20, 2023 2616 Views

Quantitative Determination of Cholesterol Hydroxylase Specificities by GC–MS/MS in Living Mammalian Cells
Hodaka Saito [...] Yoshio Yamauchi
Jan 20, 2024 2380 Views
Abstract
Glycerophospholipids consist of a glycerophosphate backbone to which are esterified two acyl chains and a polar head group. The head group (e.g., choline, ethanolamine, serine or inositol) defines the glycerophospholipid class, while the acyl chains together with the head group define the glycerophospholipid molecular species. Stable heavy isotope (e.g., deuterium)-labeled head group precursors added to the culture medium incorporate efficiently into glycerophospholipids of mammalian cells, which allows one to determine the rates of synthesis, acyl chain remodeling or turnover of the individual glycerophospholipids using mass spectrometry. This protocol describes how to study the metabolism of the major mammalian glycerophospholipids i.e., phosphatidylcholines, phosphatidylethanolamines, phosphatidylserines and phosphatidylinositols with this approach.
Keywords: GlycerophospholipidBackground
Radiolabeled precursors have been extensively used to study glycerophospholipid (GPL) metabolism in cultured cells. However, this approach has serious drawbacks. First of all, it is unfeasible to study the metabolism of all molecular species of GPLs due to the fact that it is not possible, without reverting to highly complicated and time-consuming protocols, to separate the individual molecular species from each other (Patton et al., 1982), which is obviously necessary to study their metabolism. Second, the radioisotopes needed are quite expensive. Third, for optimal labeling, the unlabeled precursors should be depleted of the medium as much as possible. Fourth, only two different precursors (labeled with 3H or 14C) can be added to the cells simultaneously and even then accurate correction for the overlap between the isotope spectra is necessary upon liquid scintillation counting of the collected fractions. Because of those handicaps, a number of studies have recently introduced an alternative approach to study GPL metabolism (e.g., Heikinheimo and Somerharju, 2002; de Kroon, 2007; Kainu et al., 2008; Postle and Hunt, 2009; Kainu et al., 2013; Hermansson et al., 2016). This approach is based on the use of stable heavy isotope-labeled precursors and mass spectrometric (MS) analysis of GPL labeling. Clearly, this approach is far more convenient and efficient as compared to the traditional methods based on the use of radiolabeled precursors, due to the following facts: (a) multiple labeled precursors can be added simultaneously to the culture medium thus providing information on several GPL classes, (b) labeled and unlabeled GPLs can be conveniently and selectively detected using head group–specific scanning modes, (c) information is obtained on the turnover of individual GPL molecular species and (d) the stable heavy isotope-labeled precursors are generally much cheaper than the radiolabeled ones and can thus be added in amounts which avoids the use of special media depleted of the corresponding unlabeled precursors.
Materials and Reagents
- Pipette tips (Thermo Fisher Scientific, Thermo ScientificTM, catalog numbers: 9400327 , 9401255 , 9401410 )
- 35 or 60 mm cell culture dishes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 153066 or 150288 )
- 12 ml screw-cap glass tubes (Kimble Chase Life Science and Research Products, catalog number: 45066A-16100 )
- Pasteur pipettes (BRAND, catalog number: 747720 )
- 12 x 32 mm screw-neck vials with caps (WATERS, catalog number: 186002640 )
- 11.5 x 75 mm test tubes (VWR, catalog number: ZZ130296772 )
- Cell scrapers and lifters (VWR, catalog numbers: 734-2603 and 734-2602 )
- HeLa cells or other cultured mammalian cells
- Phosphate buffered saline (PBS) (Merck Millipore, catalog number: 524650 )
- Methanol (VWR, catalog number: 83638.320 )
- Internal standards needed for mass spectrometric analyses:
- Di-20:1-phosphatidylcholine (PC) (Avanti Polar Lipids, catalog number: 850396 )
- Di-20:1-phosphatidylethanolamine (PE; synthesized in-house from corresponding PC) (Käkelä et al., 2003)
- Di-20:1-phosphatidylserine (PS; synthesized from corresponding PC) (Käkelä et al., 2003)
- Di-16:0-phosphatidylinositol (PI) (Avanti Polar Lipids, catalog number: 850141 )
- Chloroform (Merck Millipore, catalog number: 102445 )
- Acetonitrile, LC-MS grade (Fisher Scientific, catalog number: A/0638/17X )
- Isopropanol, OptimaTM LC/MS grade (Fisher Scientific, catalog number: A461-212 )
- Ammonium formate (Sigma-Aldrich, catalog number: 70221 )
- Ammonia solution 25% Suprapur (Merck Millipore, catalog number: 105428 )
- Acetic acid, glacial (Fisher Scientific, catalog number: 10171460 )
- Deuterium-labeled choline (D9-choline chloride) (C/D/N ISOTOPES, catalog number: D-2142 )
- Deuterium-labeled ethanolamine (D4-ethanolamine) (Cambridge Isotope Laboratories, catalog number: DLM-552-1 )
- Deuterium-labeled serine (D3-L-serine) (Cambridge Isotope Laboratories, catalog number: DLM-1073-1 )
- Deuterium-labeled inositol (D6-myo-inositol) (C/D/N ISOTOPES, catalog number: D-3019 )
- Hydroxylamine (Sigma-Aldrich, catalog number: 55460 )
- Dulbecco’s modified Eagle medium high glucose (DMEM) (Thermo Fisher Scientific, GibcoTM, catalog number: 41965039 ) or another medium appropriate for the cell line of interest
Note: We grew HeLa cells in DMEM containing 10% fetal calf serum and 100 U/ml penicillin and 100 U/ml streptomycin. - Choline chloride (Sigma-Aldrich, catalog number: C1879 )
- Ethanolamine (Merck Millipore, catalog number: 800849 )
- Myo-inositol (Sigma-Aldrich, catalog number: I5125 )
- L-serine (Sigma-Aldrich, catalog number: S4500 )
- Fetal bovine serum (FBS) (Sigma-Aldrich, catalog number: F7524 )
- Penicillin-streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- Milli-Q H2O (Elga Stat Maxima Reverse Osmosis Water Purifier)
- Nitrogen gas (Aga, Industrial gases, 99.95%)
- Labeling medium (see Recipes)
- Chase medium (see Recipes)
Equipment
- Pipettes (Thermo Fisher Scientific, Thermo ScientificTM, catalog numbers: 4600170 , 4600240 and 4600250 )
- Microliter syringe (Hamilton, model: 705 N, catalog number: 80565 )
- Vortex mixer (VWR, catalog number: 444-1372 )
- Centrifuge (Thermo Fisher Scientific, model: HeraeusTM MegafugeTM 1.0 )
- Fume hood
- Sample Concentrator with nitrogen evaporation (Cole-Parmer, Techne, catalog number: FSC400D )
- Mass spectrometer or analysis of the samples by a service provider
Note: We used Quattro Micro and Quattro Premier triple-quadrupole mass spectrometers (WATERS, model: Quattro Premier Mass Spectrometers ) - Acquity FTN Ultra Performance Liquid Chromatography instrument (WATERS, model: ACQUITY UPLC H-Class System )
- 1.0 x 150 mm Acquity BEH C18 column (WATERS, catalog number: 186002347 )
Software
- MassLynx 4.1 and QuanLynx software (WATERS)
- LIMSA software (Haimi et al., 2009)
Procedure
- Metabolic labeling of newly synthesized phospholipids
To determine the time course of incorporation of deuterium labeled head groups into glycerophospholipids you need several dishes with adherent cells. The cells from a dish are collected at suitable times e.g., after 0, 1, 2, 4, 8, 12, 24, and 48 h of incubation with the labeled head group precursors. - Grow cells until dishes are 70-80% confluent. Adherent cells lines like HeLa, CHO or BHK-21 cells are suitable.
- Wash cells once with 1 ml of PBS to remove detached cells.
- Add 1 or 1.5 ml of the labeling medium (see Recipes) per 35 or 60 mm dish and incubate the cells for up to 48 h.
- Collect cells from a dish at suitable times. Wash the dish thrice with 1 ml of PBS and collect the cells by scraping in 2 ml of ice-cold H2O and move the cells to 12 ml screw-cap tubes.
- Vortex briefly, suspend cells with a pipette and collect e.g., 50 µl aliquots of the cell suspension for protein determination, if necessary.
- To the remaining suspension add 3 ml of methanol and vortex.
- Store at -20 °C until ready for extraction of the lipids.
- Pulse-chase experiments
To determine the turn-over of glycerophospholipids first incubate the cells with the labeled precursors for up to 24 h, wash the cells with PBS and then incubate them in a medium containing the corresponding unlabeled head group precursors. During this chase period, collect cells from a dish at e.g., 0, 1, 2, 4, 8, and 24 h of incubation to determine the rate of turnover. - Label the newly synthesized phospholipids as described in steps 1a-1c for 4-24 h.
- Wash thrice with cell culture medium to remove the labeled precursors.
- Add 1 or 1.5 ml of the chase medium to each dish and incubate for up to 48 h.
- Collect cells (see steps 1d-1g) first at a 1-2 h interval and then with longer intervals.
- Extraction of lipids
- Add a cocktail of internal GPL standard consisting of di 20:1-PC, di 20:1-PE, di 20:1-PS and di 16:0-PI (10, 5, 2, 2 nmol per 100 nmol of total glycerophospholipid, respectively) for mass spectrometric analyses and then extract the lipids (see Notes). We use the Folch method of lipid extraction (Folch et al., 1957) except that no added salts are included.
- Add 6 ml of chloroform, add the caps and turn the tubes upside down and back again rapidly 40-50 times.
- Centrifuge at 3,000 x g for 10 min.
- Transfer the lower phase with a glass Pasteur pipette to a new 12 ml screw-cap tube, add 4 ml of theoretical upper phase containing chloroform/methanol/H2O (3:48:47, v/v) and mix as described in step 3b.
- Centrifuge as in step 3c and collect the lower phase.
- Add 0.5 ml of methanol, vortex, and evaporate to dryness under nitrogen stream using mild heating (< 40 °C).
- Reconstitute extracted lipids in 20-40 µl of methanol/chloroform (4:1, v/v) in 12 x 32 mm screw-neck vials for LC-MS analysis, or in 250 µl of methanol/chloroform (2:1, v/v) in 11.5 x 75 mm test tubes for MS/MS analysis and store at -20 °C.
- Mass spectrometric analysis
Note: It can be carried out using either LC-MS or MS/MS using direct sample infusion as indicated below. - Liquid chromatography-mass spectrometry
GPL molecular species were analyzed using an LC-MS system consisting of an Acquity FTN Ultra Performance Liquid Chromatography instrument (WATERS) equipped with a 1.0 x 150 mm Acquity BEH C18 column connected to Quattro Micro or Premier triple-quadrupole mass spectrometer (WATERS). The column was eluted with a gradient of isopropanol/acetonitrile (90:10) into acetonitrile/H2O (60:40) each containing 10 mM ammonium formate and 1% NH4OH (Kainu et al., 2013). The flow rate was 0.13 ml/min. Individual lipid species were detected using Multiple Reaction Monitoring (MRM) as described (Kainu et al., 2013). Mass spectrometer was operated in the positive ion mode and the precursor ions were [M + H]+ for PC, PE, PS and PI. The product ions for the unlabeled species were m/z 184 (PC), 141 (PE), 185 (PS) and 260 (PI) and for the deuterium-labeled species m/z 193, 145, 188 and 266, respectively. Typical m/z transitions are listed in Table 1. Water soluble GPL metabolites were also analyzed with LC-MS as described (Hermansson et al., 2016). Data were collected using the MassLynx 4.1 software (WATERS). - MS/MS analysis with direct sample infusion
This method does not require LC instrument because the different GPLs in the crude lipid extract can be selectively detected based on their head group–specific fragmentation when the sample is infused to the mass spectrometer with a glass microsyringe at a rate of 10-20 µl/min. The unlabeled PC and PI species are selectively detected by scanning for precursors of m/z +184 and -241, respectively, while the unlabeled PE and PS species are detected by monitoring for a neutral loss (NL) of 141 or 87, respectively. The labeled PC, PI or PE and PS species are selectively detected by scanning for the precursors of 193 (PC) or 247 (PI) or neutral loss of 145 (PE) or 90 (PS).
Table 1. MRM precursor and product ions used for select GPL molecular species in LC-MS analysis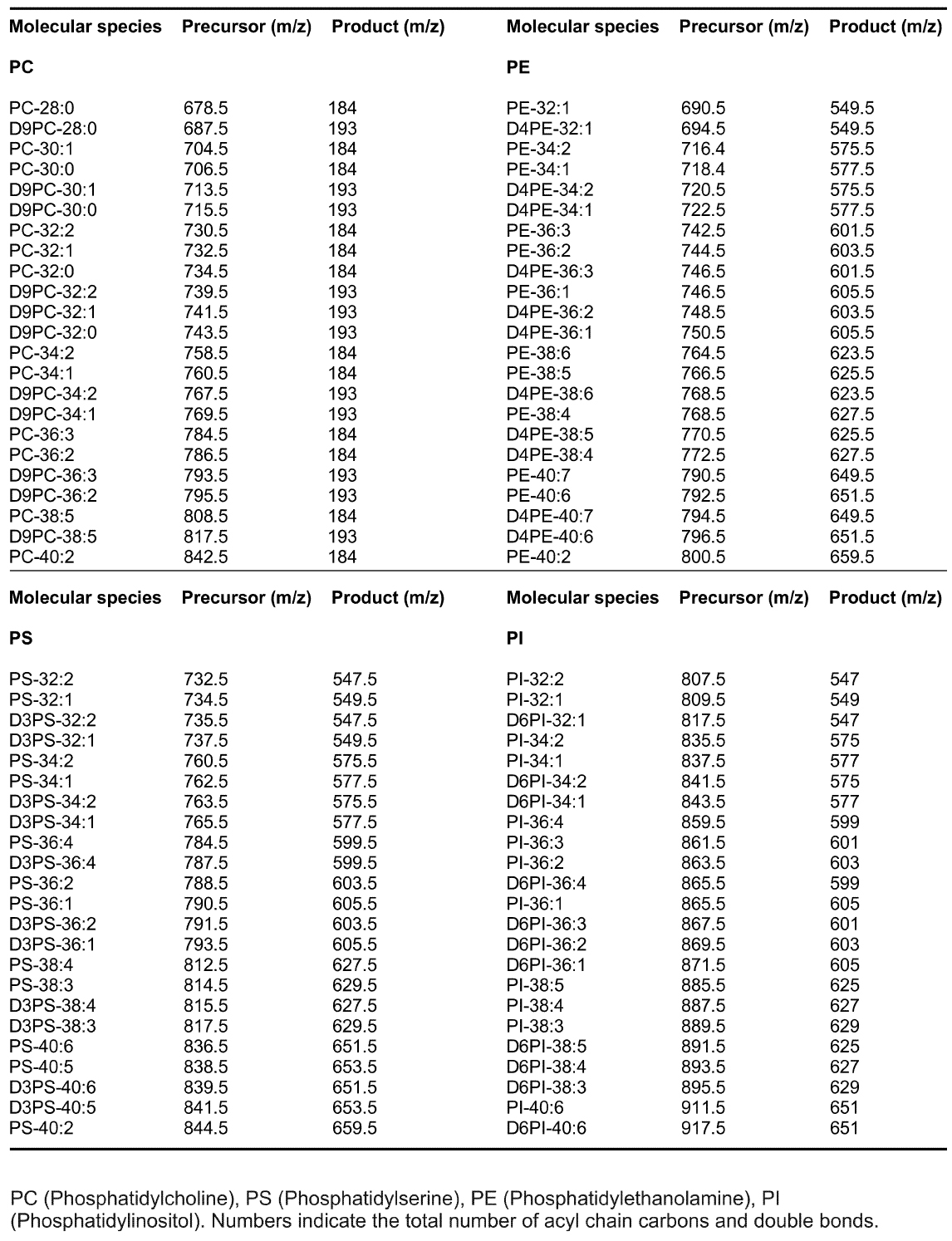
Data analysis
- The analysis method depends on the method used for data acquisition. In case of the LC-MS method, the labeled and unlabeled lipids (and their metabolites) were quantified using the QuanLynx software (WATERS).
- When the MS/MS method with direct infusion was used, the head group–specific MS/MS spectral data were copied to Excel and then the LIMSA add-in was run. This provides the concentrations of the individual GPL species based on the included internal standards (Haimi et al., 2009). When needed, GPL concentration vs. time was plotted and the turnover rate was obtained by curve fitting with the Origin software (OriginLab, Northampton, MA).
- Typical results
Figure 1 shows the labeling of one representative PC species from D9-choline added to the growth medium of HeLa cells.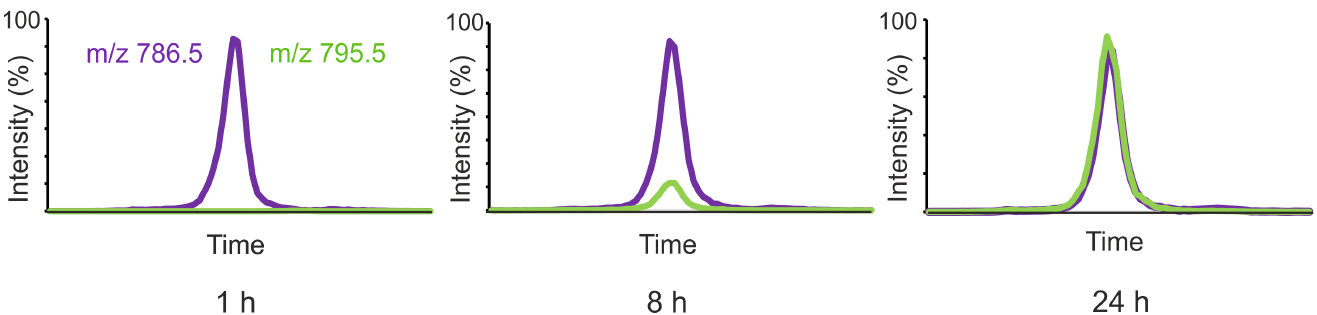
Figure 1. HeLa cells were incubated in a medium containing D9-choline for 1, 8 or 24 h and the cellular lipids were extracted and the labeled and unlabeled PC species were analyzed by LC-MS using MRM detection. Unlabeled 36:2-PC (m/z 786.5) is shown in purple and the corresponding labeled PC species (m/z 795.5) in green.
Notes
Many GPL standards are available from e.g., Avanti Polar Lipids (Alabaster, AL).
For most accurate quantification, a heavy isotope -labeled internal standard for each lipid species to be analyzed should be used (Krautbauer et al., 2016; Wang et al., 2016), but this is rarely done due to lack of such commercial standards. However, in the present approach labeling is determined from the intensity of a labeled GPL species relative to that of the corresponding unlabeled one and thus no internal standards are necessary.
Recipes
- Labeling medium
- Adjust pH of the D4-ethanolamine solution to 7.0 with acetic acid and mix it with the other deuterium-labeled precursors and hydroxylamine in H2O to obtain the precursor stock solution (typically 100x). Store at -20 °C
Note: pH of ethanolamine is high and thus it needs to be neutralized with acetic acid to avoid alkalization of the labeling and chase media. - Prepare the labeling medium by adding stock solution in the growth medium to obtain the following concentrations: D9-choline 100 µg/ml, D4-ethanolamine 100 µg/ml, D3-L-serine 300 µg/ml, D6-myo-inositol 100 µg/ml, and 1 mM hydroxylamine
Note: Hydroxylamine is an inhibitor of PS decarboxylase that prevents decarboxylation of phosphatidylserine (PS) to phosphatidylethanolamine (PE) in the cells. The presence of hydroxylamine is necessary to avoid the MS peaks due to the labeled PE species formed via the CDP-ethanolamine pathway from overlapping with the peaks of PE species formed via decarboxylation of labeled PS. However, if the PE species formed from PS via decarboxylation are of interest, hydroxylamine as well as labeled ethanolamine must be omitted from the medium to avoid such overlap. - Chase medium
DMEM containing unlabeled choline (500 µg/ml), ethanolamine (500 µg/ml), L-serine (1,000 µg/ml), myo-inositol (500 µg/ml) and 1 mM hydroxylamine was stored at 4 °C and used on the same day
Acknowledgments
This protocol was used in the following publications: Kainu et al., 2013; Hermansson et al., 2016; Kainu et al., 2008. This study was supported by grants from Finnish Academy and Sigrid Juselius Foundation to P.S.
References
- de Kroon, A. I. (2007). Metabolism of phosphatidylcholine and its implications for lipid acyl chain composition in Saccharomyces cerevisiae. Biochim Biophys Acta 1771(3): 343-352.
- Folch, J., Lees, M. and Sloane Stanley, G. H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226(1): 497-509.
- Haimi, P., Chaithanya, K., Kainu, V., Hermansson, M. and Somerharju, P. (2009). Instrument-independent software tools for the analysis of MS-MS and LC-MS lipidomics data. Methods Mol Biol 580: 285-294.
- Heikinheimo, L. and Somerharju, P. (2002). Translocation of phosphatidylthreonine and -serine to mitochondria diminishes exponentially with increasing molecular hydrophobicity. Traffic 3(5): 367-377.
- Hermansson, M., Hanninen, S., Hokynar, K. and Somerharju, P. (2016). The PNPLA-family phospholipases involved in glycerophospholipid homeostasis of HeLa cells. Biochim Biophys Acta 1861(9 Pt A): 1058-1065.
- Kainu, V., Hermansson, M., Hanninen, S., Hokynar, K. and Somerharju, P. (2013). Import of phosphatidylserine to and export of phosphatidylethanolamine molecular species from mitochondria. Biochim Biophys Acta 1831(2): 429-437.
- Kainu, V., Hermansson, M. and Somerharju, P. (2008). Electrospray ionization mass spectrometry and exogenous heavy isotope-labeled lipid species provide detailed information on aminophospholipid acyl chain remodeling. J Biol Chem 283(6): 3676-3687.
- Käkelä, R., Somerharju, P. and Tyynela, J. (2003). Analysis of phospholipid molecular species in brains from patients with infantile and juvenile neuronal-ceroid lipofuscinosis using liquid chromatography-electrospray ionization mass spectrometry. J Neurochem 84(5): 1051-1065.
- Koivusalo, M., Haimi, P., Heikinheimo, L., Kostiainen, R. and Somerharju, P. (2001). Quantitative determination of phospholipid compositions by ESI-MS: effects of acyl chain length, unsaturation, and lipid concentration on instrument response. J Lipid Res 42(4): 663-672.
- Krautbauer, S., Buchler, C. and Liebisch, G. (2016). Relevance in the use of appropriate internal standards for accurate quantification using LC-MS/MS: Tauro-conjugated bile acids as an example. Anal Chem 88: 10957-10961.
- Patton, G. M., Fasulo, J. M. and Robins, S. J. (1982). Separation of phospholipids and individual molecular species of phospholipids by high-performance liquid chromatography. J Lipid Res 23(1): 190-196.
- Postle, A. D. and Hunt, A. N. (2009). Dynamic lipidomics with stable isotope labelling. J Chromatogr B Analyt Technol Biomed Life Sci 877(26): 2716-2721.
- Wang, M., Wang, C. and Han, X. (2016). Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry-what, how and why? Mass Spectrom Rev.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hänninen, S., Somerharju, P. and Hermansson, M. (2017). Metabolic Heavy Isotope Labeling to Study Glycerophospholipid Homeostasis of Cultured Cells. Bio-protocol 7(9): e2268. DOI: 10.21769/BioProtoc.2268.
Category
Biochemistry > Lipid > Lipid measurement
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









