- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Mating Based Split-ubiquitin Assay for Detection of Protein Interactions
Published: Vol 7, Iss 9, May 5, 2017 DOI: 10.21769/BioProtoc.2258 Views: 17315
Reviewed by: Arsalan DaudiRia SircarTimothy Notton

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Split-luciferase Complementation Imaging Assay to Study Protein-protein Interactions in Nicotiana benthamiana
Liping Wang [...] Rosa Lozano-Durán
Dec 5, 2021 12064 Views

In vitro Auto- and Substrate-Ubiquitination Assays
Hye Lin Park [...] Gyeong Mee Yoon
Apr 5, 2022 3524 Views

Assessing Metabolite Interactions With Chloroplastic Proteins via the PISA Assay
Anna Karlsson [...] Elton P. Hudson
May 5, 2025 2011 Views
Abstract
The mating based split-ubiquitin (mbSUS) assay is an alternative method to the classical yeast two-hybrid system with a number of advantages. The mbSUS assay relies on the ubiquitin-degradation pathway as a sensor for protein-protein interactions, and it is suitable for the determination of interactions between full-length proteins that are cytosolic or membrane-bound. Here we describe the mbSUS assay protocol which has been used for detecting the interaction between K+ channel and SNARE proteins (Grefen et al., 2010 and 2015; Zhang et al., 2015 and 2016)
Keywords: mbSUSBackground
Figure 1 is an overview of the mbSUS assay. The ubiquitin moiety is split into two halves, and the N-terminal half is mutated (NubG) to avoid reassembly. The C-terminal half of the ubiquitin moiety (Cub) is linked with the transcription reporter complex PLV (Protein A-LexA-VP16). The fusion of two proteins (X and Y) to NubG and CubPLV, respectively, yields a system for protein-protein interaction analysis. After transformation, the yeast strain THY.AP5 contains the NubG-X fusion protein, while yeast strain THY.AP4 contains the Y-CubPLV fusion protein. After mating, in the diploid yeast, if proteins X and Y interact with each other, a functional ubiquitin will be reassembled which leads to cleavage of the PLV. The released transcription protein complex PLV switches on reporter genes (ADE2, HIS3) and allows yeast growth on selective media.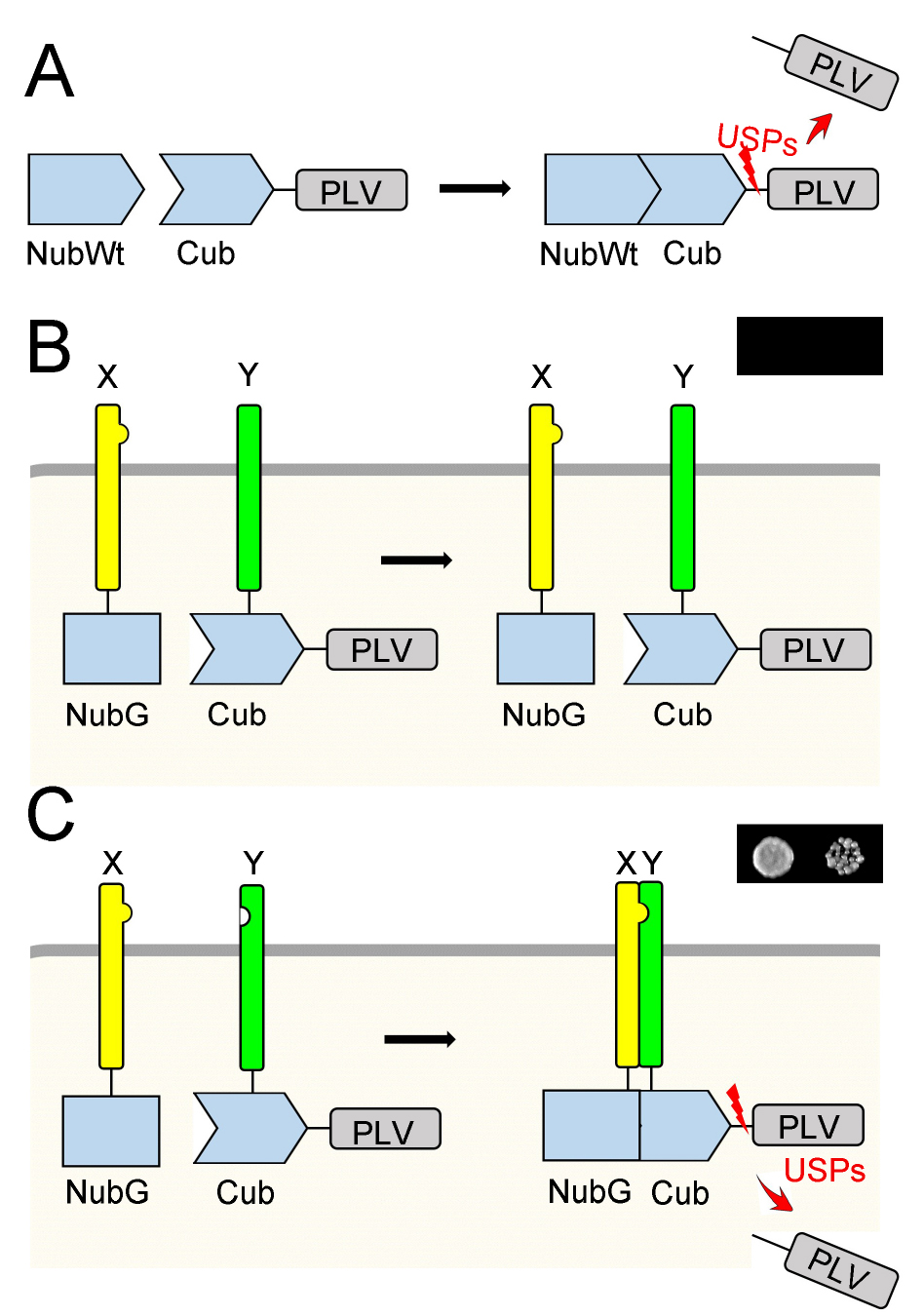
Figure 1. The split-ubiquitin system. A. The Ubiquitin is split into two halves, the N-terminal wild type half (NubWt; NubI) and the C-terminal half (Cub). Reassembly of NubWt and Cub leads to the release of the transcription reporter complex Protein A-LexA-VP16 (PLV) by ubiquitin-specific proteases (USPs). B. Mutating the isoleucine at position 13 of the N-terminal half to glycine yields the NubG protein, which inhibits the spontaneous reassembly of ubiquitin. In diploid yeast, NubG-X and Y-CubPLV fusion proteins are produced. If X and Y do not interact, no functional ubiquitin is formed, and the yeast cannot grow on selective media as shown in a small image on top-right. C. If X interacts with Y, then NubG and Cub can reassemble as a functional ubiquitin, which leads to the release of PLV. The PLV activates the reporter genes (ADE2, HIS3) synthesising ADE and HIS, which allows yeast growth on the selective media without these chemicals as shown in a small image on top-right.
Materials and Reagents
- Sterile toothpick
- Round bottom polystyrene tubes, 14 ml (purchased as sterile) (Corning, catalog number: 352051 )
- Blue screw cap tubes, 50 ml (purchased as sterile; Cellstar)
- Screw cap tubes, 2 ml (purchased as sterile; SARSTEDT)
- PCR tubes, 0.2 ml, flat cap (Sterilized by autoclaving) (STARLAB INTERNATIONAL, catalog number: I1402-8100 )
- Nitrocellulose transfer membranes, BioTraceTM NT (Pall, catalog number: 66485 )
- 1.5 ml screw cap tube
- Minisart® syringe filter, non-pyrogenic, 0.2 µm (Sartorius)
- Syringe filter w/0.45 µm polyethersulfone membrane (VWR, catalog number: 28145-503 )
- Blotting paper (VWR, Blotting pad 707)
- Petri dishes, 55 mm (purchased as sterile) (Greiner Bio One International, catalog number: 628102 )
- Square Petri dishes, 120 x 120 mm (purchased as sterile) (Greiner Bio One International, catalog number: 688102 )
- Filter tips 10, 200, 1,000 μl (Biosphere® Plus, for micropipettes, SARSTEDT)
- Saccharomyces cerevisiae yeast strains used in the mbSUS assay (Table 1)
Table 1. Yeast strains used for mbSUS assayName Genotype Function Reference THY.AP4 MATa;
ade2-, his3-, leu2-, trp1-,
ura3-;
lexA::ADE2,
lexA::HIS3, lexA::lacZBait: carrying the Y-
CubPLV in vector
pMetYC-Dest(Obrdlik et al., 2004) THY.AP5 MATα;
URA3;
ade2-, his3-, leu2-, trp1-Prey: carrying the
NubG-X in vector
pNX35-Dest or NubWt(Obrdlik et al., 2004) - Destination vectors used in the mbSUS assay (Table 2)
Table 2. The destination vectors used for mbSUS assayVector name Promotor Gateway site Function Reference pMetYC-Dest met25 attR1, attR2 Fusion protein with C-terminal
CubPLV as bait; synthesis of
LEU in yeast(Grefen et al., 2009) pNX35-Dest ADH1 attR1, attR2 Fusion protein with N-terminal
NubG as prey; synthesis of
TRP in yeast(Grefen and Blatt, 2012) - Donor vector: pDONR207 vector (Thermo Fisher Scientific) or other donor vectors contain attP1, attP2 Gateway cassette
- Gateway® clonase enzyme:
- BP Clonase®II Enzyme Mix (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11789020 )
- LR Clonase®II Enzyme Mix (Thermo Fisher Scientific, InvitrogenTM, catalog number: 11791020 )
- Protein ladder (Pageruler Plus Prestained Protein Ladder, Thermo Fisher Scientific)
- Western blot signal detection kit (SuperSignal West Dura Chemiluminescent Substrate) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 37071 )
- Antibody solutions
- Primary: anti-HA (1:20,000, Anti-HA high-affinity Rat monoclonal antibody) (Roche Diagnostics, catalog number: 11867423001 ) or anti-VP16 (1:20,000, Anti-VP16 tag antibody in Rabbit) (Abcam, catalog number: ab4808 )
- Secondary: anti-rabbit HRP (1:20,000, goat anti-rabbit IgG-HRP) (Thermo Fisher Scientific, InvitrogenTM, catalog number: G-21234 ) or anti-rat HRP (1:20,000, Rabbit anti-Rat IgG H&L [HRP]) (Abcam, catalog number: ab6734 )
- Methionine
- Glycerol (Fisher Scientific, catalog number: 10795711 )
- Peptone (FormediumTM, catalog number: PEP02 )
- Glucose (Fisher Scientific, catalog number: 10141520 )
- Yeast extract (FormediumTM, catalog number: YEA02 )
- Oxoid agar (Agar No.1) (Oxoid, catalog number: LP0011 )
- Potassium hydroxide (KOH) (Sigma-Aldrich, catalog number: 60377 )
- Lithium acetate dihydrate (Sigma-Aldrich, catalog number: L4158 )
- De-ionized water
- Acetic acid (Sigma-Aldrich, catalog number: 1.00063 )
- Salmon sperm DNA (ssDNA) (Sigma-Aldrich, catalog number: 31149 )
- Polyethylene Glycol 3350 (Spectrum Chemical Manufacturing, catalog number: Po125-500gm )
- YNB without ammonium sulphate, without potassium (MP Biomedicals, catalog number: 114029622 )
- Potassium dihydrogen orthophosphate (Fisher Scientific, catalog number: 10783611 )
- CSM-ADE-HIS-LEU-MET-TRP-URA (powder) (MP Biomedicals, catalog number: 114560222 )
- Agar
- Adenine sulphate (Sigma-Aldrich, catalog number: A3159 )
- Uracil (Sigma-Aldrich, catalog number: U1128 )
- L-leucine (Sigma-Aldrich, catalog number: L8912 )
- L-tryptophane (Sigma-Aldrich, catalog number: T4196 )
- L-histidine (Sigma-Aldrich, catalog number: H3911 )
- L-methionine (Sigma-Aldrich, catalog number: M5308 )
- Sodium dodecyl sulfate (SDS) (VWR, catalog number: 442444H )
- Urea (Sigma-Aldrich, catalog number: U5378 )
- DL-dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 43817 or 43815 )
Note: The product “ 43817 ” has been discontinued. - Acrylamide (Sigma-Aldrich, catalog number: A8887 )
- Bromophenol blue
- Ammonium persulfate (Fisher Scientific, catalog number: 10020020 )
- TEMED (Sigma-Aldrich, catalog number: T9281 )
- Ammonium sulphate (VWR, catalog number: 21333.296 )
- Tris (Fisher Scientific, catalog number: BP152-1 )
- Glycine (Fisher Scientific, catalog number: 10070150 )
- Methanol (Sigma-Aldrich, catalog number: 34860 )
- Ponceau S (Sigma-Aldrich, catalog number: P3504 )
- Sodium chloride (VWR, catalog number: 27810.295 )
- Hydrochloric acid (HCl) (Sigma-Aldrich, catalog number: H1758 )
- Tween® 20 (Sigma-Aldrich, catalog number: P1379 )
- Milk powder (Marvel, Iceland, UK)
- YPD media (see Recipes)
- LiAc solution (see Recipes)
- ssDNA solution (see Recipes)
- PEG solution (see Recipes)
- CSM-minimal media (see Recipes)
- Reagents for auxotrophy selection (see Recipes)
- SCM-auxotrophy selection media (see Recipes)
- ‘Lyse & Load’ buffer (see Recipes)
- 10% SDS-PAGE separation gel (see Recipes)
- 5% SDS-PAGE stacking gel (see Recipes)
- Bjerrums buffer (see Recipes)
- Ponceau S solution (see Recipes)
- TBS solution and TBS-Tween solution (see Recipes)
- Running buffer (see Recipes)
- Blocking solution (see Recipes)
Equipment
- Incubator shaker for yeast growth in liquid culture (Sartorius, model: BS1 )
- Glass flask, 500 ml (sterilized by autoclaving) (Corning, PYREX®, catalog number: 4980-500 )
- High speed refrigerated microcentrifuge (Eppendorf, model: 5417 R )
- Refrigerated centrifuge for large volume liquid samples (Thermo Fisher Scientific, Thermo ScientificTM, model: SorvallTM LegendTM RT )
- PCR machine (VWR, PEQLAB, model: peqSTAR 96 Universal Gradient )
- Pipette
- Incubator for yeast growth on solid media plates (Gallenkamp, model: Ipr150 )
- Ultrasonic disintegrator (MSE, model: Soniprep 150 MSS150.CX , 9.5 mm probe)
- Biophotometer (Eppendorf, model: Biophotometer Plus 6132 )
- SDS-PAGE gel electrophoresis and blotting boxes (Bio-Rad Laboratories, models: Power PAC 1000 and Mini-PROTEAN Tetra Cell )
- Shaker for Western blot (Stuart, model: Stuart STR9 Gyro rocker )
- Semi-dry blotting system (VWR, PEQLAB, model: PerfectBlueTM Semi-Dry Blotter )
- Western blot imaging platform (Vilber, model: Fusion spectra )
- Vortex (Scientific Industries, model: Vortex-Genie 2 )
- Imaging system
- Autoclave
Procedure
- Molecular biology
Gateway® Technology (Thermo Fisher Scientific, Germany) is used to create plasmids for the yeast mbSUS assay following the manufacturer’s instructions. The unique attB1 and attB2 sites (B1 5’-3’: ggggacaagtttgtacaaaaaagcaggct; B2 5’-3’: ggggaccactttgtacaagaaagctgggt) are included in the forward and reverse primers as overhanging sequences, respectively. To fuse the PCR product in the proper reading frame with an N-terminal tag, the forward primer must include two additional nucleotides. Depending on the destination vector, a stop codon is included in the reverse primer or provided on the vector itself.
Subsequent PCR products are inserted by BP reactions into the pDONR207 vector to yield Entry clones that are verified by DNA sequencing. Gateway Destination clones, pNX35-Dest for prey (Grefen and Blatt, 2012) and pMetYC-Dest for bait (Grefen et al., 2009), are generated by using LR Clonase® II in LR reactions according to the manufacturer’s instructions. - Yeast transformation
The yeast transformation is performed under sterile condition. Prepare fresh competent yeast for each transformation.- Pick THY.AP4 and AP5 yeast from long-term stock by a sterile toothpick or pick one colony of yeast from a stock plate (see Note 7) and inoculate 5 ml YPD in 14 ml sterile round bottom tubes and incubate in a shaker overnight at 200 rpm, 28 °C.
- Inoculate 45 ml of YPD with the 5 ml of overnight culture in autoclaved 500 ml flasks and incubate with shaking at 200 rpm, 28 °C for 3-5 h until OD600 0.6-0.8.
- Harvest cells by centrifugation (10 min at 2,000 x g, 4 °C) in 50 ml blue screw cap tubes and discard the supernatant.
- Wash cells with 20 ml of sterile water, centrifuge again, and discard the supernatant.
- Resuspend the cells with 1.8 ml of 0.1 M LiAc and transfer to a 2 ml Eppendorf tube, spin down (30 sec at 2,000 x g, 4 °C), and discard the supernatant.
- Add an appropriate amount of 0.1 M LiAc (multiply the number of transformations by 20 µl) and incubate at room temperature for 30 min.
- Meanwhile, prepare sterilized PCR tubes with 10 µl of ssDNA and 5 µl of plasmid DNA (Destination clones, > 100 ng/μl) for each transformation.
- Prepare a master mix: mix 70 µl of PEG solution, 10 µl of 1 M LiAc, and 20 µl of competent yeast (from step B6) for each transformation.
- Mix the master mix with the DNA mixture from step B7.
- In a PCR machine, incubate at 30 °C for 30 min, then gently mix the mixture by pipette and heat shock cells at 43 °C for 30 min.
- Spin down the yeast at 2,000 x g for 1 min, and discard the supernatant by pipetting it carefully off the pelleted cells.
- Wash the cells with 100 µl of sterile water, spin down, and discard the supernatant.
- Resuspend cells in 50 µl of sterile water and plate on the appropriate selective media.
- THY.AP4 carrying bait vector plates on CSM-LM solid medium.
- THY.AP5 carrying prey vector plates on CSM-TUM solid medium.
- Incubate for 48-72 h at 28 °C. At least 100 colonies are expected in each plate.
- Pick THY.AP4 and AP5 yeast from long-term stock by a sterile toothpick or pick one colony of yeast from a stock plate (see Note 7) and inoculate 5 ml YPD in 14 ml sterile round bottom tubes and incubate in a shaker overnight at 200 rpm, 28 °C.
- mbSUS assay
The mbSUS assay is performed under sterile conditions.- Pick 5-15 colonies of transformed THY.AP5 yeast carrying the prey construct and inoculate in 3 ml CSM-TUM media. Grow overnight in the incubator shaker at 200 rpm, 28 °C.
- Pick 5-15 colonies of transformed THY.AP4 yeast carrying the bait construct and inoculate in 3 ml CSM-LM media. Grow overnight in the incubator shaker at 200 rpm, 28 °C.
- Reserve 750 µl from each bait and prey yeast culture to store as a stock culture (see Note for yeast stock preparation).
- Harvest yeast cells by centrifugation at 2,000 x g for 1 min at room temperature and resuspend the pellets in YPD media (for each mating 20 µl of YPD media should be calculated).
- Mix 20 µl of each yeast (carrying bait) and yeast (carrying prey) in sterilized PCR tube and immediately drop 4 µl of each mated yeast onto YPD plate. Incubate overnight at 28 °C.
- Transfer the mated yeast from the YPD plate to a CSM-LTUM plate with a sterile toothpick (Figure 2A). Grow overnight at 28 °C.
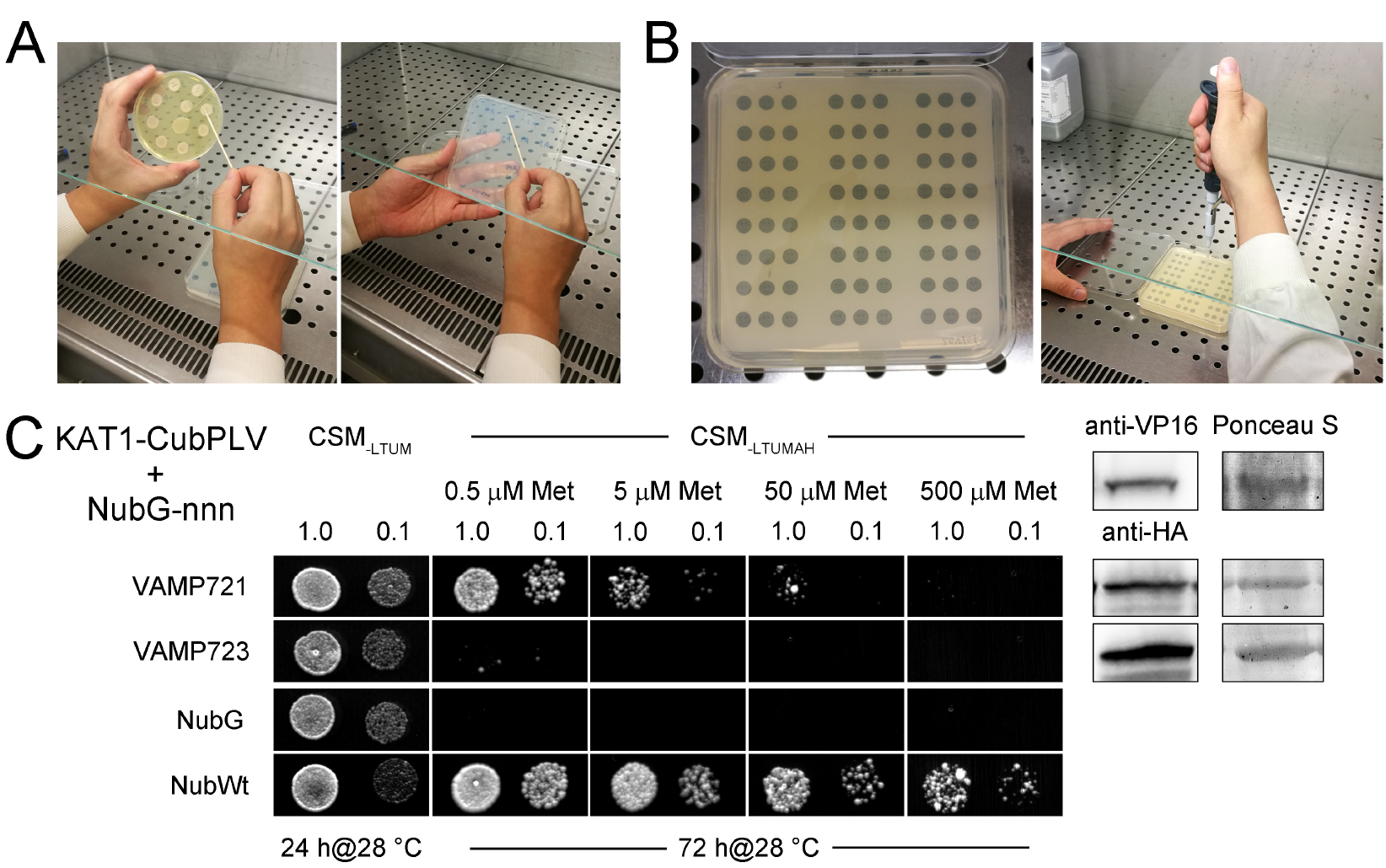
Figure 2. Example of a mbSUS assay. A. Yeast is transferred from the YPD plate to a CSM-LTUM plate with a sterile toothpick. B. Yeast is dropped on the CSM-LTUMAH plate with black spots on paper. C. Diploid yeast expressing KAT1-CubPLV as bait with NubG-X fusions of VAMP721, VAMP723, and the controls (negative: NubG; positive: NubWt) as prey were spotted onto different media as indicated. CSM-LTUM was used to verify the presence of both bait and prey vectors. The addition of different concentration of methionine (Met) to CSM-LTUMAH was used to verify interaction when K+ channel-CubPLV expression was suppressed. Diploid yeast was dropped at 1.0 and 0.1 OD600 in each case. Incubation time was 24 h for CSM-LTUM plate and 72 h for CSM-LTUMAH plates. Western blot analysis of the diploid yeast used in mbSUS was carried out with commercial HA antibody for the VAMP fusions and VP16 antibody for the K+ channel fusions. Ponceau S stains were used as blotting/loading control (right). - Pick the mated yeast colony of each sample from the CSM-LTUM plate and inoculate in 3 ml liquid CSM-LTUM media, grow overnight in the incubator shaker at 200 rpm, 28 °C.
- Inoculate 1 ml of the mated cells into 3 ml of fresh CSM-LTUM media and grow at 200 rpm, 28 °C for 4-6 h for later Western blot analysis.
- Determine OD600 of yeast cells from step C7.
- Harvest 100 µl of remaining cells from step C7 by centrifugation at 2,000 x g, 1 min at room temperature and dilute them to OD600 1.0 and 0.1 respectively in sterile water.
- Apply 4 µl drops of each dilution on CSM-LTUM and CSM-LTUMAH plates with different methionine concentrations (0.5, 5, 50, and 500 µM; Figure 2B), grow them at 28 °C. Check for growth after 1 day and 3 days growth on CSM-LTUM plate and CSM-LTUMAH plate respectively. Figure 2C shows an example of a mbSUS assay which indicates an interaction between VAMP721 and KAT1 K+ channel, and no interaction between VAMP723 and KAT1 (Zhang et al., 2015). Diploid yeast containing NubWt as prey serves as the positive control and yeast containing NubG as prey is the negative control. In a good mbSUS assay, the sample of negative control should show weak or no yeast growth on CSM-LTUMAH plates, while the yeast grows well in the positive control sample (Figure 2C).
- Pick 5-15 colonies of transformed THY.AP5 yeast carrying the prey construct and inoculate in 3 ml CSM-TUM media. Grow overnight in the incubator shaker at 200 rpm, 28 °C.
- Yeast protein extraction for western blot analysis
- Determine the OD600 of the yeast culture.
- Harvest 2 ml of the culture and resuspend in ‘Lyse & Load’ buffer at a final OD600 of 100 for each sample. The volume of ‘Lyse & Load’ buffer used is calculated like this: e.g., the 1:10 diluted culture measurement yields an OD600 of 0.5, which is equivalent to an OD600 of 5 for 1 ml of culture. Harvest 2 ml of the culture yields OD600 of 10. Then resuspend them in 0.1 ml of ‘Lyse & Load’ buffer can get a final OD600 of 100 in the sample.
- Lyse the yeast with an ultrasonic disintegrator (amplitude setting 10 for 20 sec, keep samples on ice).
- Incubate for 10 min at 65 °C.
- Vortex for 10 sec, spin down the lysate at full speed for 30 sec and transfer the supernatant to a fresh tube. Then sample can be used for Western blot analysis.
- Determine the OD600 of the yeast culture.
- Western blot analysis
- Load protein ladder and 10-15 μl of each sample on an SDS-PAGE gel.
- Run gel with appropriate conditions and transfer proteins to a nitrocellulose membrane (e.g., by semidry transfer).
- Block the membrane for 60 min in the blocking buffer at room temperature.
- Transfer the membrane to the primary antibody solution; incubate for at least 3 h at room temperature or overnight at 4 °C.
- Wash 3 times with 1x TBS-Tween, 10 min each wash.
- Transfer the membrane into the secondary antibody solution; incubate for at least 3 h at room temperature or overnight at 4 °C.
- Wash 3 times with 1x TBS-Tween, 10 min each wash.
- Incubate the membrane with the chemiluminescent substrate for 5 min in darkness.
- Develop the membrane and take images with an imaging system.
- Wash membrane several times with distilled water to remove the chemiluminescent substrate.
- Incubate the membrane in the Ponceau S solution for 5 min.
- Wash the membrane several times with distilled water until the red bands are clearly visible.
- Scan Ponceau S image as blotting/loading control.
- Load protein ladder and 10-15 μl of each sample on an SDS-PAGE gel.
Notes
- When using proteins fusions, the C-terminus of the native protein with the Y-CubPLV fusion should be located in the cytosol and the NubG-X fusion of the prey should also be available in the cytosol for effective interactions to take place. Reassembly of Cub and Nub then leads to cleavage of the PLV moiety and allows this transcription factor to move into the nucleus.
- Macromolecules smaller than 40 kDa can passively diffuse through the nuclear pore, and some larger particles are also able to pass through the large diameter of the pore (Marfori et al., 2011). So the bait protein should normally be anchored to a membrane. Soluble bait fusions could otherwise move into the nuclear and activate the reporter genes independent of any interaction and PLV cleavage. Preys fusions need not be membrane anchored.
- Expression of fusion proteins in the bait vector pMetYC-Dest is under control of the met25 promoter which suppresses expression in the presence of methionine in the culture media (Grefen et al., 2009). However, our experience of mbSUS assay shows that the yeast growth on media without external methionine is slower than the yeast on media adding 5 μM methionine. To overcome this problem, it is recommended to add 0.5 μM methionine to the media. Higher concentrations, up to 1 mM Met, are used to test for specificity in interaction by reducing the bait expression.
- Before yeast dropping, it is recommended to allow media plates to dry. Allow the surface of the plate to dry for 30 min under sterile conditions. The dry surface of the solid media in plate helps to keep the yeast-spot round.
- When dropping yeast, it is recommended to print a dropping scheme with black spots on paper and stick it under the plate, as shown in Figure 2B left.
- For Western blot analysis, we recommend that the yeast is not cultured overnight. Add 3 ml fresh media to a 1 ml yeast culture from the previous overnight growth and allow the culture to grow for a further 4-6 h before harvesting. This approach avoids the possibility of senescent yeast in the culture which may introduce degraded protein and lead to poor Western blot results.
- To prepare yeast stock for long-term storage, pipette 750 µl of fresh overnight yeast culture (from step C1 or C2) and add 250 µl of sterile 60% glycerol to a 1.5 ml screw cap tube, tightly close the cap and mix before freezing at -80 °C. It is recommended to refresh glycerol stocks every 6 months and avoid repeated freeze-thaw cycles of the glycerol stock. To recover yeast cells from a glycerol stock, pick frozen yeast cells using a sterile toothpick, drop and spread onto the appropriate selective media solid plate, then incubate at 28 °C for 24-48 h. Yeast colonies on selective media are ready to use for mbSUS assay, and the yeast colonies can be stored at 4 °C for up to 1 month. To prepare a yeast stock for short-term storage, pipette 5-10 µl of fresh overnight yeast culture (from step C1 or C2) or pick frozen yeast cells from glycerol stock; and drop cells on appropriate selective solid media. Culture yeast for 24-48 h at 28 °C, then store at 4 °C for up to1 month.
Recipes
- YPD media
2% peptone
2% glucose
1% yeast extract
2% agar
Adjust pH to 6.0 with 1 N KOH
Sterilize by autoclaving and store at 4 °C - LiAc solution (1 M or 0.1 M)
Dissolve lithium acetate in de-ionized water. Adjust the pH to 7.5 with acetic acid, sterilize by filtration (Minisart® Syringe Filter) and store at room temperature - ssDNA solution
Dissolve 10 mg/ml ssDNA in de-ionized water, sterilize by filtration (Minisart® Syringe Filter) and boil for 5 min following cooling on ice before use. Store at -20 °C - PEG solution
Dissolve PEG 3350 in de-ionized water to a final concentration of 50% (w/v), sterilize by filtration (0.45 µm polyethersulfone membrane) and avoid water loss during storage as this significantly decreases the transformation efficiency. Store at room temperature - CSM-minimal media
0.7 g/L YNB (without ammonium sulphate, without potassium)
5 g/L ammonium sulphate
1 g/L potassium dihydrogen orthophosphate
20 g/L glucose
0.55 g/L of CSM-dropout mix, adjust pH to 6.0 with fresh prepared KOH
Add 20 g/L agar if solid media is needed. The YNB and CSM-dropout-mixes contained all amino acids apart from the desired selective one(s). Sterilize by autoclaving and store at 4 °C - Reagents for auxotrophy selection
Prepared by dissolving the following each chemical in 100 ml of water, sterilized by filtration, and stored in darkness at 4 °C:
ADE: 0.4 g of adenine sulphate (add 5 ml per liter media)
URA: 0.4 g of uracil (add 5 ml per liter media)
LEU: 2.0 g of L-leucine (add 5 ml per liter media)
TRP: 0.4 g of L-tryptophane (add 5 ml per liter media)
HIS: 0.4 g of L-histidine (add 5 ml per liter media)
MET: 1.5 g of L-methionine (equals a 0.1 M stock solution, add appropriate amount to obtain 0.5, 5, 50, and 500 μM final concentrations) - CSM-auxotrophy selection media
The CSM-auxotrophy selection media are prepared by adding reagents for auxotrophy selection (see Recipe 6) into the CSM-minimal media after autoclaving and cooling down to around 50 °C as shown below (Table 3):
Table 3. The SC-auxotrophy selection media used for mbSUS assayMedia Reagents for auxotrophy selection Comment CSM-TUM media ADE, HIS, LEU Transformation of Nub-clones in THY.AP5. CSM-LM media ADE, HIS, TRP, URA Transformation of Cub-clones in THY.AP4. CSM-LTUM media ADE, HIS Yeast mating check CSM-LTUMAH media with Met MET Yeast growth check; different methionine
concentrations can help to increase the signal-to-noise ratio - ‘Lyse & Load’ buffer
50 mM Tris-HCl (pH 6.8)
4% SDS
8 M urea
30% glycerol
0.1 M DTT
0.005% bromophenol blue
Store at -20 °C - 10% SDS-PAGE separation gel (50 ml)
17 ml acrylamide (30%)
12.5 ml 1.5 M Tris-HCl (pH 8.8)
0.5 ml 10% SDS
0.5 ml 10% ammonium persulfate
0.05 ml TEMED
Add ddH2O to 50 ml - 5% SDS-PAGE stacking gel (50 ml)
8.3 ml acrylamide (30%)
6.3 ml 1 M Tris-HCl (pH 6.8)
0.5 ml 10% SDS
0.5 ml 10% (NH4)2S2O8
0.05 ml TEMED
Add ddH2O to 50 ml - Bjerrums buffer
6.1 g/L Tris
2.4 g/L glycine
20% methanol
Store at 4 °C - Ponceau S solution
2 g/L Ponceau S in 5 % acetic acid
Store at room temperature - TBS solution and TBS-Tween solution
8 g/L NaCl
0.2 g/L KCl
3 g/L Tris
Adjust pH to 7.4 with HCl
Store at room temperature
Add Tween® 20 to a final concentration 0.1% to make TBS-Tween - Running buffer
3 g/L Tris
14.4 g/L glycine
1.5 g/L SDS
Store at room temperature - Blocking solution
5% milk powder in TBS-Tween solution
Acknowledgments
This protocol was adapted and modified from a previous publication (Grefen et al., 2009). We thank Prof. Michael Blatt for his advice and support in preparing this article. We thank Dr Emily Larson for her useful suggestions. This work was supported by a Royal Thai Government Scholarship to WH (ST4718), a Chinese Scholarship Council studentship to BZ, and by funding from BBSRC grants BB/I024496/1, BB/K015893/1, BB/L001276/1, BB/M01133X/1, BB/M001601/1, and BB/L019205/1 to Prof. Blatt.
References
- Grefen, C. and Blatt, M. R. (2012). Do calcineurin B-like proteins interact independently of the serine threonine kinase CIPK23 with the K+ channel AKT1? Lessons learned from a menage a trois. Plant Physiol 159(3): 915-919.
- Grefen, C., Chen, Z., Honsbein, A., Donald, N., Hills, A. and Blatt, M. R. (2010). A novel motif essential for SNARE interaction with the K+ channel KC1 and channel gating in Arabidopsis. Plant Cell 22(9): 3076-3092.
- Grefen, C., Karnik, R., Larson, E., Lefoulon, C., Wang, Y., Waghmare, S., Zhang, B., Hills, A. and Blatt, M. R. (2015). A vesicle-trafficking protein commandeers Kv channel voltage sensors for voltage-dependent secretion. Nat Plants 1: 15108.
- Grefen, C., Obrdlik, P. and Harter, K. (2009). The determination of protein-protein interactions by the mating-based split-ubiquitin system (mbSUS). Methods Mol Biol 479: 217-233.
- Marfori, M., Mynott, A., Ellis, J. J., Mehdi, A. M., Saunders, N. F., Curmi, P. M., Forwood, J. K., Boden, M. and Kobe, B. (2011). Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim Biophys Acta 1813(9): 1562-1577.
- Obrdlik, P., El-Bakkoury, M., Hamacher, T., Cappellaro, C., Vilarino, C., Fleischer, C., Ellerbrok, H., Kamuzinzi, R., Ledent, V., Blaudez, D., Sanders, D., Revuelta, J.L., Boles, E., André, B., and Frommer, W.B. (2004). K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci USA 101: 12242-12247.
- Zhang, B., Karnik, R., Waghmare, S., Donald, N. A. and Blatt, M. R. (2016). VAMP721 conformations unmask an extended motif for K+ channel binding and gating control. Plant Physiol.
- Zhang, B., Karnik, R., Wang, Y., Wallmeroth, N., Blatt, M. R. and Grefen, C. (2015). The Arabidopsis R-SNARE VAMP721 interacts with KAT1 and KC1 K+channels to moderate K+current at the plasma membrane. Plant Cell 27(6): 1697-1717.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Horaruang, W. and Zhang, B. (2017). Mating Based Split-ubiquitin Assay for Detection of Protein Interactions. Bio-protocol 7(9): e2258. DOI: 10.21769/BioProtoc.2258.
Category
Plant Science > Plant biochemistry > Protein > Interaction
Biochemistry > Protein > Interaction > Protein-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










