- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Incubation of Cyanobacteria under Dark, Anaerobic Conditions and Quantification of the Excreted Organic Acids by HPLC
Published: Vol 7, Iss 9, May 5, 2017 DOI: 10.21769/BioProtoc.2257 Views: 9129
Reviewed by: Neelanjan BoseAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

β-lactamase (Bla) Reporter-based System to Study Flagellar Type 3 Secretion in Salmonella
Fabienne F. V. Chevance and Kelly T. Hughes
Jun 20, 2023 1776 Views

Determination of Poly(3-hydroxybutyrate) Content in Cyanobacterium Synechocystis sp. PCC 6803 Using Acid Hydrolysis Followed by High-performance Liquid Chromatography
Janine Kaewbai-ngam [...] Tanakarn Monshupanee
Aug 20, 2023 1828 Views

An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
Avisek Mahapa [...] Dipankar Chatterji
Dec 20, 2024 1791 Views
Abstract
Succinate and lactate are commodity chemicals used for producing bioplastics. Recently, it was found that such organic acids are excreted from cells of the unicellular cyanobacterium Synechocystis sp. PCC 6803 under dark, anaerobic conditions. To conduct the dark, anaerobic incubation, cells were concentrated within a vial that was then sealed with a butyl rubber cap, following which N2 gas was introduced into the vial. The organic acids produced were quantified by high-performance liquid chromatography via post-labeling with bromothymol blue as a pH indicator. After separation by ion-exclusion chromatography, the organic acids were identified by comparing their retention time with that of standard solutions. These procedures allow researchers to quantify the organic acids produced by microorganisms, contributing to knowledge about the biology and biotechnology of cyanobacteria.
Keywords: Anaerobic conditionBackground
Cyanobacteria are a group of bacteria that perform oxygenic photosynthesis. Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803) is a non-nitrogen-fixing, unicellular cyanobacterium that is commonly used for basic and applied research studies. Synechocystis 6803 cells are able to excrete organic acids, such as succinate and lactate, under both dark and anaerobic conditions (Osanai et al., 2015). Genetic manipulation and modification of the incubation conditions, such as addition of potassium or NaHCO3, have succeeded in increasing the levels of organic acids excreted from Synechocystis 6803 cells (Osanai et al., 2015; Hasunuma et al., 2016; Iijima et al., 2016; Ueda et al., 2016). Since succinate and lactate are commodity chemicals used to make various materials, such as bioplastics, their bio-based production is a desirable way to reduce the environmental burden. Herein, we describe the methods of Synechocystis 6803 incubation under dark, anaerobic conditions, and quantification of the organic acids by HPLC via post-labeling with bromothymol blue.
Materials and Reagents
- 50 ml disposable polypropylene tubes
- Test tubes for cultivation (Iwaki, catalog number: TEST30NP )
- 20 ml headspace vials (GL Sciences, catalog number: 1030-46026 )
- 22 G x 1″ needle (Terumo Medical, catalog number: NN-2225R )
- 22 G x 1 ½″ needle (Terumo, catalog number: NN-2238S )
- Aluminum foil
- SupraPure hydrophilic PVDF syringe filters (Recenttec, catalog number: R7-PVDF033S022I )
- Target DP vials (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: C4000-1 )
- Screw caps (GL Sciences, catalog number: 1030-45261 )
- 300 µl target polyspring inserts (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: C4010-S630 )
- Disposable syringes (Terumo, catalog number: SS-10LZ )
- Open-top-style aluminum caps with butyl rubber septa (Systech, catalog number: 98552 )
- MF-Millipore membrane filters (Merck Millipore, catalog number: HAWP03700 )
- Cyanobacterium Synechocystis sp. PCC 6803
- Ammonium chloride (NH4Cl) (Wako Pure Chemical Industries, catalog number: 017-02995 )
- 60% perchloric acid (HClO4) (Wako Pure Chemical Industries, catalog number: 160-05755 )
- Trichloroacetic acid
- Succinate
- Lactate
- Citric acid (Nacalai Tesque, catalog number: 09109-85 )
- Acetate
- Ferric ammonium citrate
- Na2EDTA (Nacalai Tesque, catalog number: 15111-45 )
- Sodium nitrate (NaNO3) (Wako Pure Chemical Industries, catalog number: 195-02545 )
- Potassium phosphate dibasic (K2HPO4) (Wako Pure Chemical Industries, catalog number: 164-04295 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Wako Pure Chemical Industries, catalog number: 131-00405 )
- Calcium chloride dihydrate (CaCl2·2H2O) (Wako Pure Chemical Industries, catalog number: 031-00435 )
- Sodium carbonate (Na2CO3) (Nacalai Tesque, catalog number: 31311-25 )
- Boric acid (H3BO3) (Wako Pure Chemical Industries, catalog number: 021-02195 )
- Manganese chloride tetrahydrate (MnCl2·4H2O) (Wako Pure Chemical Industries, catalog number: 139-00722 )
- Zinc sulfate heptahydrate (ZnSO4·7H2O) (Wako Pure Chemical Industries, catalog number: 264-00402 )
- Pentahydrate copper sulphate (CuSO4·5H2O) (Wako Pure Chemical Industries, catalog number: 039-04412 )
- Sodium molybdate dehydrate (Na2MoO4·2H2O) (Wako Pure Chemical Industries, catalog number: 196-02472 )
- Sulfuric acid (H2SO4) (Wako Pure Chemical Industries, catalog number: 199-15995 )
- Cobalt dinitrate hexahydrate, Co(NO3)2·5H2O (Wako Pure Chemical Industries, catalog number: 031-03752 )
- HEPES (Dojindo Molecular Technologies, catalog number: 342-01375 )
- Potassium hydroxide (KOH) (Wako Pure Chemical Industries, catalog number: 168-21815 )
- Bromothymol blue (BTB) (Wako Pure Chemical Industries, catalog number: 027-03052 )
- Ethanol (Wako Pure Chemical Industries, catalog number: 057-00451 )
- Disodium hydrogen phosphate dodecahydrate (Na2HPO4·12H2O) (Wako Pure Chemical Industries, catalog number: 196-02835 )
- Succinic acid (Wako Pure Chemical Industries, catalog number: 190-04332 )
- DL-Lactic acid lithium salt (MP Biomedicals, catalog number: 100824 )
- Sodium acetate (Wako Pure Chemical Industries, catalog number: 190-01071 )
- Modified BG-11 liquid medium (see Recipes)
- Mobile phase (3 mM HClO4) (see Recipes)
- Reaction solution (0.2 mM BTB in 15 mM Na2HPO4) (see Recipes)
- 500 mM succinate solution (see Recipes)
- 500 mM lactate solution (see Recipes)
- 500 mM citrate solution (see Recipes)
- 500 mM acetate solution (see Recipes)
- 1 M acetate solution (see Recipes)
Equipment
- Cultivation chamber (TOMY Digital Biology, model: CLE-303 )
- A gas mixer (KOFLOC, model: RK120XM )
- An air pump (Yasunaga, model: LP-30A )
- Spectrophotometer (Shimadzu, model: UV-2700 )
- High-speed refrigerated microcentrifuge (TOMY Digital Biology, model: MX-305 )
- N2 generator (Sanyo Denshi, model: SN4-4 )
- Rotary shaker (Nissin, model: NX-20D )
- Freeze dryer (Tokyo Rikaikai, EYELA, model: FDU-2200 )
- Ultrasonic cleaning machine (SND, model: US-108 )
- Autoclave
- The HPLC system (JASCO International, model: LC-2000Plus ) was composed of the following equipment:
- Photodiode array (PDA) detector (JASCO International, model: MD-2018Plus )
- Intelligent HPLC pump (JASCO International, model: PU-2080Plus )
- Line degasser (JASCO International, model: DG-2080-53 )
- Ternary gradient unit (JASCO International, model: LG-2080-02 )
- Intelligent column oven (JASCO International, model: CO-2065Plus )
- Intelligent autosampler (JASCO International, model: AS-2057Plus )
- Ion-exclusion column (300 x 8.0 mm) (GL Sciences, model: RSpak KC-811, catalog number: 5055-13509 ) and guard column (10 x 4.6 mm) (GL Sciences, model: GPC KF-G, catalog number: 5055-13150 )
- Reaction coil unit (JASCO International, model: RU-2080-51 )
- Photodiode array (PDA) detector (JASCO International, model: MD-2018Plus )
Software
- ChromNAV software (Ver. 1.14)
Procedure
- Cultivation under light, aerobic (a) and incubation under dark, anaerobic conditions (b)
- The glucose-tolerant strain (designated as GT) of Synechocystis sp. PCC 6803, isolated by Williams (1988), is cultivated in modified BG-11 medium (see Recipes), consisting of BG-110 liquid medium (Rippka, 1988) supplemented with 5 mM NH4Cl (buffered with 20 mM HEPES-KOH, pH 7.8). In our study, the GT-I strain was used (Kanesaki et al., 2012). The GT cells are cultivated in liquid modified BG-11 medium under air containing 1% (v/v) CO2, using a gas mixer (RK120XM, KOFLOC) and an air pump (LP-30A, Yasunaga) at the flow rate of 30-50 ml/min, in a cultivation chamber (CLE-303, TOMY). The temperature is kept at 30 °C and white light is supplied continuously at 50-70 µmol photons·m-2 sec-1. Cell densities are measured at A730 using the Shimadzu UV-2700 spectrophotometer. Cells cultivated in 70 ml of modified BG-11 medium (started from A730 = 0.4) for 3 days under, light, aerobic condition. The light, aerobic cultivation affects the levels of succinate and lactate, and optimization of culture conditions is required for each laboratory.
- The cell are collected by centrifugation at 5,600 x g for 2 min and concentrated into 10 ml of HEPES buffer (20 mM HEPES-KOH, pH 7.8) or modified BG-11 to reach an A730 of 20 in a vial (Figure 1). The vial is sealed with open-top-style aluminum caps with butyl rubber septa, and two needles (Needle 22 G x 1″ and Needle 22 G x 1 ½″) are stabbed through a septum. The 22 G x 1 ½″ needle is connected to a N2 gas generator to introduce an N2 gas for 30-60 min in order to generate the anaerobic condition.
- After introduction of the N2 gas, the needles are removed and the vial is wrapped with aluminum foil. The cells are incubated at 30 °C with shaking on a rotary shaker at 160 rpm for 1-5 days. For 1 day, succinate and lactate were less excreted from the cells. Succinate and lactate levels increased during long incubation, however, the levels reached maximum at 3 or 4 days, and started to decrease at 5 days (Osanai et al., 2015).

Figure 1. A 20 ml vial for anaerobic incubation. Cells were concentrated into 10 ml of buffer or medium to reach an A730 of 20, and the vial was then sealed with a butyl rubber cap. Two syringes were inserted into the cap, and N2 gas was introduced through one of two syringes for 30-60 min. After introduction of the N2 gas, the syringes were removed and the vial was wrapped in aluminum foil to maintain a dark condition. - Sample preparation for HPLC analysis
- After incubation, the aluminum cap with butyl rubber is removed with an opener. The cell cultures are centrifuged at 5,600 x g for 2 min. Using a SupraPure sterile syringe filter, the supernatants are filtered to remove debris.
- Then, 1 ml of supernatant is transferred into a 2 ml plastic tube. The supernatants are placed in a flask and dried at -80 °C with an FDU-2200 freeze dryer for 1 day. The freeze-dried samples are resolved with 100 µl of 3 mM perchloric acid (HClO4) (since HClO4 is highly corrosive, it should be handled with care) and centrifuged at 5,600 x g for 2 min.
- Then, transfer 100 µl of the supernatant into a Target DP vial using a glass insert. Set it in the autosampler for HPLC analysis.
- In case of samples containing large amount of proteins, freeze-dried samples should be resolved in 97 µl of 3 mM HClO4 and 3 µl of 100% trichloroacetic acid. Mix thoroughly and then centrifuge the samples at 20,400 x g for 10 min. Use the supernatant for HPLC analysis.
- To prepare standards, mix 100 μl of 500 mM succinate solution, 160 μl of 500 mM lactate solution, 100 μl of 500 mM citrate solution, 200 μl of 1 M acetate solution (see Recipes), and 440 μl of dH2O to make up a total 1 ml of organic acids standard. This solution was named STD1. The final concentration of each organic acid in STD1 is 80 mM lactate, 50 mM succinate, 50 mM citrate, and 200 mM acetate.
- Dilute STD1 to make a range of concentrations for drawing the analytical curves.
- HPLC analysis
Quantification of the organic acids is performed by HPLC using post-labeling with BTB (Figure 2). Use 3 mM HClO4 solution as the mobile phase and 0.2 mM BTB in 15 mM Na2HPO4 solution as the reaction solution. The mobile phase and reaction solution are degassed for approximately 20 min using the ultrasonic cleaning machine and the aspirator. HPLC measurement is performed with the following conditions: - Intelligent HPLC pump (Mobile phase): the flow rate is set at 0.7 ml/min. The pump is connected to the line degasser and the ternary gradient unit, and further connected to the intelligent column oven. Before sample injection, equilibrate the columns with mobile phase at same flow rate for one hour.
- Intelligent HPLC pump (Reaction solution): the flow rate is set at 1.2 ml/min.
- Intelligent column oven: the column temperature is 60 °C.
- Intelligent autosampler: the sampler temperature is 4 °C and the injection volume is 20 µl. To clean up the injection needle, use HClO4 solution. Flush three times with HClO4 before sample injection.
- Reaction coil unit was used for mixing mobile phase including samples and BTB solution.
- PDA: single beam photometer with deuterium (D2) and halogen (W) lamps; single data analysis is performed for 42 min using scans in the wavelength range of 200-900 nm. Before starting the HPLC analysis, wait for approximately 60 min until the flow is stable (baseline without drift and noise).
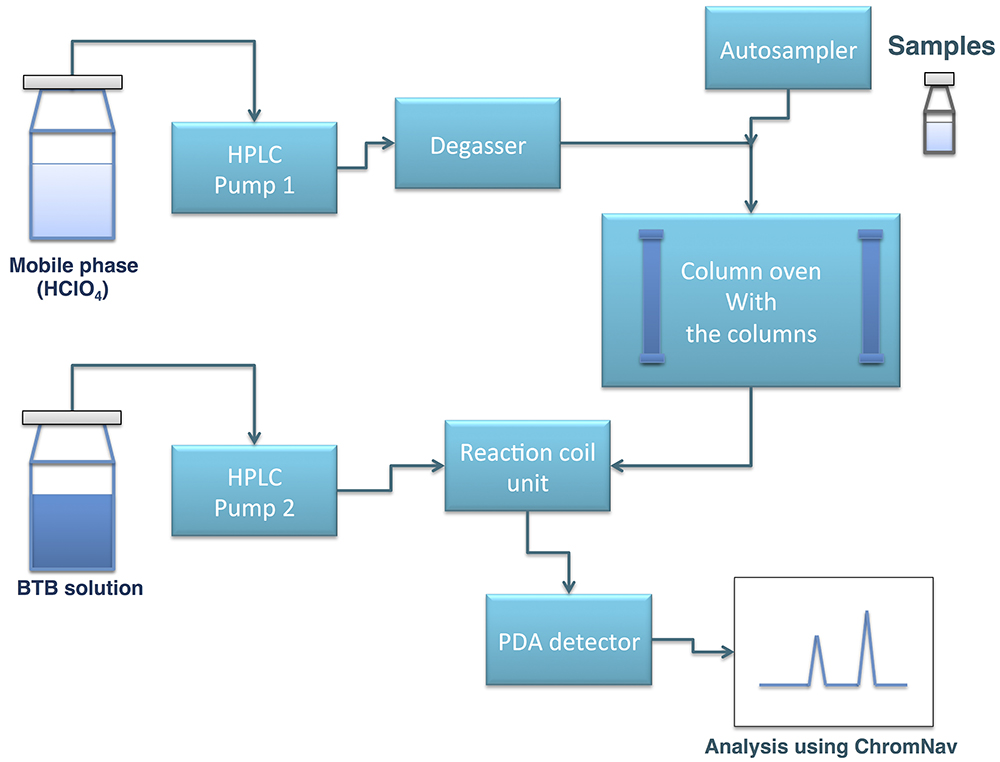
Figure 2. Schematic diagram of HPLC analysis with bromothymol blue post-labeling. Two HPLC pumps are used for mobile phase and BTB solution, respectively. Samples are injected with autosampler and divided by size exclusion columns, followed by mixed with BTB solution. Organic acids are detected by PDA detector and analyzed by ChromNAV software.
Data analysis
Typical chromatograms measured by HPLC with BTB post-labeling are shown in Figure 3. Organic acid peaks are quantified using the absorbance at 445 nm. Use the ChromNAV program to quantify the peak areas of each organic acid. Draw standard curves for each organic acid by marking with a dot of known concentration samples. Calculate the concentrations of organic acids in the samples by using the following standard curves (Figure 4).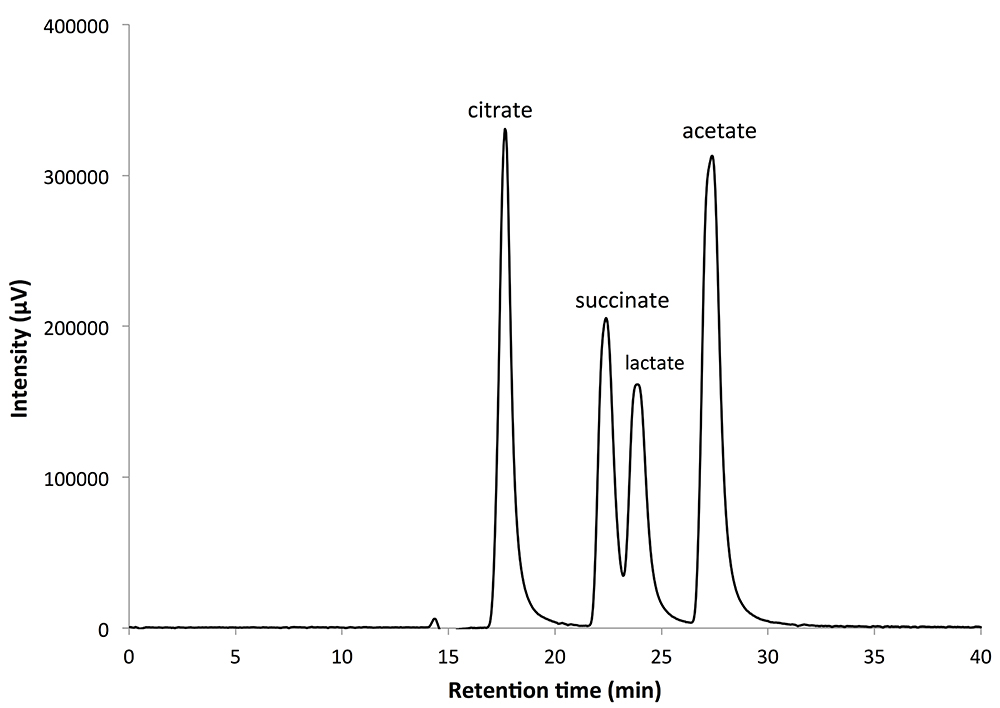
Figure 3. Typical chromatograms measured by HPLC with bromothymol blue post-labeling. The peak intensity was calculated from the absorbance at wavelength 445 nm. The retention times for each organic acid were as follows: citrate 17.55 min, succinate 22.36 min, lactate 23.80 min, and acetate 27.34 min.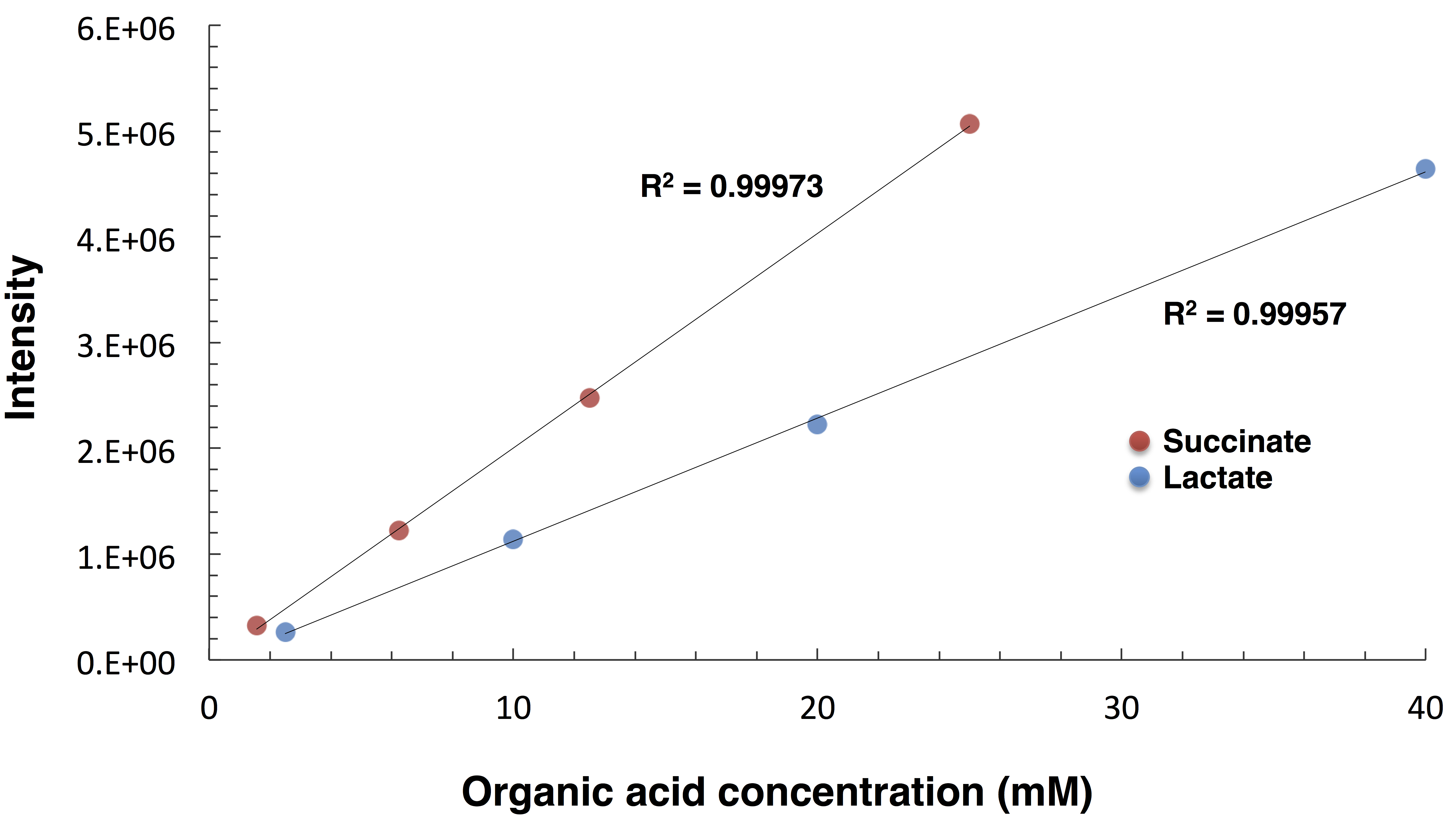
Figure 4. Standard curves for succinate (orange) and lactate (blue). The intensity values were calculated from the chromatograms by integrating the peak areas of each organic acid.
Notes
In case of samples containing large amount of proteins, freeze-dried samples should be resolved in 97 µl of 3 mM HClO4 and 3 µl of 100% trichloroacetic acid. Mix thoroughly and then centrifuge the samples at 20,400 x g for 10 min. Use the supernatant for HPLC analysis.
Recipes
- Modified BG-11 liquid medium (store at room temperature)
- Solution 1 (store at 4 °C)
Weigh 0.6 g citric acid, 0.6 g ferric ammonium citrate, and 0.1 g Na2EDTA, and add distilled water (dH2O) to 200 ml - Solution 2 (store at 4 °C)
Weigh 30 g NaNO3, 0.78 g K2HPO4, and 1.5 g MgSO4·7H2O, and add dH2O to 1 L - Solution 3 (store at 4 °C)
Weigh 3.8 g CaCl2·2H2O and add dH2O to 200 ml - Solution 4 (store at 4 °C)
Weigh 4 g Na2CO3 and add dH2O to 200 ml - Solution 5 (store at 4 °C)
Weigh 2.86 g H3BO3, 1.81 g MnCl2·4H2O, 0.22 g ZnSO4·7H2O, 0.08 g CuSO4·5H2O, 0.021 g Na2MoO4, and 50 μl H2SO4, and add dH2O to 1 L (1*)
Weigh 0.0494 g Co(NO3)2·5H2O and add to (1*) - 1 M HEPES-KOH, pH 7.8 (store at 4 °C)
Weigh 119.1 g HEPES and add dH2O to approximately 400 ml
Adjust pH to 7.8 with 6 N KOH and add dH2O to 500 ml
Autoclave (121 °C for 15 min) and store at 4 °C - K2HPO4 solution (store at 4 °C)
Weigh 0.39 g K2HPO4 and add dH2O to 200 ml - MgSO4·7H2O solution (store at 4 °C)
Weigh 0.75 g MgSO4·7H2O and add dH2O to 200 ml
Mix 20 ml 1 M HEPES-KOH (pH 7.8), 50 ml Solution 2, 2 ml Solution 3, 1 ml Solution 4, and 1 ml Solution 5, and add dH2O to 1 L
Autoclave the mixture and store at room temperature
Before use, add 2 ml Solution 1 to 1 L mixture - Mobile phase (3 mM HClO4)
Weigh 0.5 g 60% HClO4 and add dH2O to 1 L
Filter with the MF-Millipore membrane filter - Reaction solution (0.2 mM BTB in 15 mM Na2HPO4)
Weigh 125 mg BTB and add to 10 ml ethanol (1)
Weigh 5.3 g Na2HPO4·12H2O and add to dH2O (2)
Mix (1) and (2), and add dH2O to 1 L
Filter the mixture with the MF-Millipore membrane filter - 500 mM succinate solution
Weigh 590.45 mg succinate and add to 10 ml dH2O using a measuring flask - 500 mM lactate solution
Weigh 480.05 mg DL-lactate lithium salt and add to 10 ml dH2O using a measuring flask - 500 mM citrate solution
Weigh 960.6 mg citrate and add dH2O to 10 ml using a measuring flask - 1 M acetate solution
Weigh 820.3 mg sodium acetate and add dH2O to 10 ml using a measuring flask
Acknowledgments
This protocol was adapted from our previous studies published by Ueda et al. (2016) and Iijima et al. (2016). This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology, Japan, by a grant to T.O. from ALCA (Project name ‘Production of cyanobacterial succinate by the genetic engineering of transcriptional regulators and circadian clocks’) from the Japan Science and Technology Agency, and by JSPS KAKENHI Grant-in-Aid for Scientific Research on Innovative Areas (Grant No. 16H06559).
References
- Hasunuma, T., Matsuda, M. and Kondo, A. (2016). Improved sugar-free succinate production by Synechocystis sp. PCC 6803 following identification of the limiting steps in glycogen catabolism. Metab Eng Commun 3: 130-141.
- Iijima, H., Shirai, T., Okamoto, M., Pinto, F., Tamagnini, P., Hasunuma, T., Kondo, A., Hirai, M., T. and Osanai, T. (2016). Metabolomics-based analysis revealing the alteration of primary carbon metabolism by the genetic manipulation of a hydrogenase HoxH in Synechocystis sp. PCC 6803. Algal Res 18: 305-313.
- Kanesaki, Y., Shiwa, Y., Tajima, N., Suzuki, M., Watanabe, S., Sato, N., Ikeuchi, M. and Yoshikawa, H. (2012). Identification of substrain-specific mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803. DNA Res 19(1): 67-79.
- Osanai, T., Shirai, T., Iijima, H., Nakaya, Y., Okamoto, M., Kondo, A. and Hirai, M. Y. (2015). Genetic manipulation of a metabolic enzyme and a transcriptional regulator increasing succinate excretion from unicellular cyanobacterium. Front Microbiol 6: 1064.
- Rippka, R. (1988). Isolation and purification of cyanobacteria. Methods Enzymol 167: 3-27.
- Ueda, S., Kawamura, Y., Iijima, H., Nakajima, M., Shirai, T., Okamoto, M., Kondo, A., Hirai, M. Y. and Osanai, T. (2016). Anionic metabolite biosynthesis enhanced by potassium under dark, anaerobic conditions in cyanobacteria. Sci Rep 6: 32354.
- Williams, J. G. K. (1988). Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol. 167: 766-778.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yasuda, C., Iijima, H., Sukigara, H. and Osanai, T. (2017). Incubation of Cyanobacteria under Dark, Anaerobic Conditions and Quantification of the Excreted Organic Acids by HPLC. Bio-protocol 7(9): e2257. DOI: 10.21769/BioProtoc.2257.
Category
Microbiology > Microbial metabolism > Carbohydrate
Microbiology > Microbial biochemistry > Other compound
Biochemistry > Carbohydrate > Lactate
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










