- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Immunoprecipitation of Cell Surface Proteins from Gram-negative Bacteria
(*contributed equally to this work) Published: Vol 7, Iss 9, May 5, 2017 DOI: 10.21769/BioProtoc.2250 Views: 9533
Reviewed by: Valentine V TrotterChristian RothAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1584 Views

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1927 Views

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2171 Views
Abstract
The meningococcus (Neisseria meningitidis) remains an important threat to human healthworldwide. This Gram-negative bacterium causes elevated disabilities and mortality in infectedindividuals. Despite several available vaccines, currently there is no universal vaccine against allcirculating meningococcal strains (Vogel et al., 2013). Herein, we describe a new protocol that iscapable of identifying only cell surface exposed proteins that play a role in immunity, providing thisresearch field with a more straightforward approach to identify novel vaccine targets. Even though N. meningitidis is used as a model in the protocol herein described, this protocol can be used for anyGram-negative bacteria provided modifications and optimizations are carried out to adapt it to differentbacterial and disease characteristics (e.g., membrane fragility, growth methods, serum antibody levels,etc.).
Keywords: Gram-negativeBackground
Attempts to develop novel vaccines against N. meningitidis often rely on 2D SDS-PAGE (two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis) and Western blotfollowed by MS (mass spectrometry) (Wheeler et al., 2007). However, such approach employs wholecell lysate, identifying a plethora of proteins that do not have vaccine potential (Mendum et al., 2009).We therefore aimed at developing a method capable of identifying only cell surface exposed proteinsthat might play important role in immunity. Briefly, our protocol consists in growing the pathogen ofinterest, immunoprecipitating surface antigens with sera of immune individuals, and identifyingimmunoprecipitated proteins by liquid chromatography-tandem mass spectrometry. We were able toidentify 23 meningococcal surface antigens using this new protocol, some of which are components ofcommercially available vaccines (Newcombe et al., 2014). We also have adapted this protocol to otherGram-negative bacteria and have obtained promising results: we identified previously describedsurface-exposed proteins, many of which have already been tested as vaccine or diagnostic testcandidates. These results show this is a robust technique that can be applied to a diverse range ofGram-negative bacteria and capable of yielding high-quality results that can be further exploited by amyriad of applications (e.g., vaccines, diagnosis, etc.).
Materials and Reagents
- Disposable Petri dishes (Cromwell Group, catalog number: STS3855002B )
- L-shaped cell spreaders (Fisher Scientific, catalog number: 14-665-231 )
- Disposable inoculating loop (Sigma-Aldrich, catalog number: I8388 )
- 1.5 ml microcentrifuge tubes (Corning, Axygen®, catalog number: MCT-150-C )
- Protein LoBind tube (Eppendorf, catalog number: 022431102 )
- 20 ml plastic universals (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 128BFS )
- Plastic sealable bags (Fisher Scientific, catalog number: 10366984)
Manufacturer: MINIGRIP, catalog number: BAJ-340-091N . - PierceTM spin columns (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 69725 )
- Disposable sterile scalpel No. 10 (WMS, catalog number: W259 )
- 10 µl Filter tips (STARLAB INTERNATIONAL, TipOne®, catalog number: S1121-3810 )
- 20 µl Filter tips (STARLAB INTERNATIONAL, TipOne®, catalog number: S1120-1810 )
- 200 µl Filter tips (STARLAB INTERNATIONAL, TipOne®, catalog number: S1120-8810 )
- 1,000 µl Filter tips (STARLAB INTERNATIONAL, TipOne®, catalog number: S1122-1830 )
- Neisseria meningitidis (strains L9153 and MC58, Public Health England)
- Phosphate-buffered saline (PBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10010001 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A9418 )
- Human serum (Sigma-Aldrich, catalog number: H6914 )
- Disease state serum (acquisition of this varies and depends on the pathogen being investigated)
- PierceTM Protein A/G UltraLinkTM Resin (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 53132 )
- SeeBlue® protein marker (Thermo Fisher Scientific, NovexTM, catalog number: LC5925 )
- Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 43815 )
- Iodoacetamide (Sigma-Aldrich, catalog number: I6125 )
- InvitrogenTM NovexTM NuPAGETM 10x sample reducing agent (Thermo Fisher Scientific, NovexTM, catalog number: NP0004 )
- InvitrogenTM NovexTM NuPAGETM 4x LDS loading buffer (Thermo Fisher Scientific, NovexTM, catalog number: NP0007 )
- InvitrogenTM NovexTM NuPAGETM 12% Bis-Tris 1 mm–10 wells (Thermo Fisher Scientific, InvitrogenTM, catalog number: NP0341BOX )
- InvitrogenTM NovexTM NuPAGETM MOPS running buffer 20x (Thermo Fisher Scientific, NovexTM, catalog number: NP000102 )
- SimplyBlueTM Safe Stain (Thermo Fisher Scientific, NovexTM, catalog number: LC6060 )
- Acetonitrile for HPLC-MS (Fisher Scientific, catalog number: 10616653 )
- Ammonium bicarbonate (Sigma-Aldrich, catalog number: 09830 )
- Trypsin Gold, mass spectrometry grade (Promega, catalog number: V5280 )
- Columbia blood agar base (Oxoid, catalog number: CM0331 )
- Horse blood, defibrinated (Oxoid, catalog number: SR0050 )
- Tris base (Roche Diagnostics, catalog number: 10708976001 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9625 )
- EDTA (Sigma-Aldrich, catalog number: E6758 )
- Triton X-100 (Sigma-Aldrich, catalog number: X100 )
- Sodium deoxycholate (Sigma-Aldrich, catalog number: 30970 )
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771 )
- Formic acid LC/MS (Fisher Scientific, catalog number: A117-50 )
- Water for HPLC-MS (Fisher Scientific, catalog number: 10777404 )
- Methanol (Sigma-Aldrich, catalog number: 179957-1L )
- Acetic acid (Sigma-Aldrich, catalog number: A6283 )
- CAB medium (see Recipes)
- PBS plus 1% BSA (see Recipes)
- Solubilization buffer (see Recipes)
- 1% SDS (see Recipes)
- 1 M dithiothreitol stock solution (see Recipes)
- 1 M Iodoacetamide (see Recipes)
- 25 mM ammonium bicarbonate (see Recipes)
- Acetonitrile (50% v/v) in 25 mM ammonium bicarbonate (see Recipes)
- Acetonitrile (50% v/v) (see Recipes)
- Solvent A for HPLC (see Recipes)
- Solvent B for HPLC (see Recipes)
- 0.1% formic acid/50% acetonitrile solution (see Recipes)
Equipment
- Gilson Pipette Pipetman Classic P2 (Gilson, catalog number: F144801 )
- Gilson Pipette Pipetman Classic P20 (Gilson, catalog number: F123600 )
- Gilson Pipette Pipetman Classic P200 (Gilson, catalog number: F123601 )
- Gilson Pipette Pipetman Classic P1000 (Gilson, catalog number: F123602 )
- Eppendorf® Micro centrifuge 5415R (Eppendorf, model: 5415 R )
- BOD low temperature refrigerated incubator (VWR, model: BOD Incubator 2005 , catalog number: 35960-056)
- CO2 incubator (LEEC, model: Culture Safe Precision 190D )
- Stuart Gyratory rocker (Cole-Parmer, Stuart, model: SSL3 )
- Vortex Mixer SA8 (Cole-Parmer, Stuart, model: SA8 )
- Ultrasonic bath (Fisher Scientific, model: FisherbrandTM S-Series , catalog number: 10611983)
- Ultimate 3000 nano HPLC system (Thermo Fisher Scientific, Thermo ScientificTM, model: UltiMateTM 3000 RSL Cnano System )
- AcclaimTM C-18 PepmapTM column (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 164569 )
- QSTAR® XL hybrid LC/MS/MS system (Applied Biosystems, model: QSTAR® XL Hybrid LC/MS/MS System )
- Eppendorf MixMateTM (Fisher Scientific, catalog number: 21-379-00)
Manufacture: Eppendorf, catalog number: 022674200 . - Eppendorf ThermoStat Plus interchangeable block heater (Eppendorf, model: ThermoStat Plus , catalog number: 022670204)
- Eppendorf VacufugeTM concentrator (Eppendorf, model: VacufugeTM Concentrator , catalog number: 022820001)
- Heraeus Tube roller Spiramix (Fisher Scientific, catalog number: 10600653)
Manufacturer: Thermo Fisher Scientific, Thermo ScientificTM, catalog number: DS507 . - Fume hood
Software
- Chromeleon v6.8 or later (Dionex, Thermo Fisher Scientific)
- Analyst QS 1.1 (Applied Biosystems)
- Mascot v2.2.04 or later (Matrix Science, London)
Procedure
- Cell surface immunoprecipitation
- Grow N. meningitidis from frozen stock: Obtain a small amount of frozen culture with a sterile 200 µl tip, inoculate Columbia agar base (CAB) plate and spread to give single colonies. Incubate overnight (~18 h) at 35 °C with 5% CO2. Collect 20-30 colonies and spread over the entire surface of a CAB plate, using a sterile loop, and incubate for 4 h at 35 °C with 5% CO2. Individual colonies will not be seen at this stage.
- Recover bacteria by adding PBS to the plate and mixing the colonies into the PBS using a sterile loop. Remove the PBS/bacteria suspension using a P1000 micropipette (1 ml per plate, use half plate for each immunoprecipitation), centrifuge for 2 min at 13,000 x g at room temperature, Wash pellet three times with 1 volume PBS plus 1% BSA. For these and subsequent wash steps, pellet cells by centrifugation for 2 min at 13,000 x g at room temperature, remove supernatant, add fresh buffer and resuspend the pellet.
Notes:- If using liquid culture instead of plates, we suggest harvesting cells at mid-log growth phase and conditions that favor the expression of virulence factors. We suggest researchers do a growth curve with their organism of interest or have good knowledge of how the organism grows in liquid media to be able to properly identify growth phases.
- Centrifugation conditions may vary according to the bacterium in question. The presence of lysed cells will increase the selection on non-surface proteins.
- Make every effort to ensure that early centrifugation steps will not disrupt cells while removing secreted proteins or components of lysed, dead cells (see Note 2).
- If using liquid culture instead of plates, we suggest harvesting cells at mid-log growth phase and conditions that favor the expression of virulence factors. We suggest researchers do a growth curve with their organism of interest or have good knowledge of how the organism grows in liquid media to be able to properly identify growth phases.
- Suspend pellet from previous step in 1 ml of serum sample (control or test [items 17 and 18 of Materials and Reagents list, respectively]) and rotate for 90 min at 4 °C.
Note: Proteins identified in the control serum samples should be disregarded in future analysis as they are likely to play no role in the immune response against the pathogen. Researchers should make sure the control sera they have is indeed negative against for the pathogen they are investigating. - Pellet the cells (13,000 x g, 5 min, RT) and wash twice with PBS plus 1% BSA.
- Resuspend cells in 1 ml solubilization buffer and incubate for 60 min at 37 °C.
- Centrifuge sample (13,000 x g, 60 min, RT), collect supernatant and incubate with protein A/GUltraLink beads (50 μl) for 18 h at 4 °C with agitation or rolling.
- Centrifuge sample (2,500 x g, 2 min, RT), collect beads.
- Wash beads five times (2,500 x g, 2 min) with 1 volume solubilization buffer.
- Elute antibody:antigen complex with 1 volume SDS (1%) and boiling (10 min).
- Add 4 volumes of water and vortex to elute proteins. Centrifuge (2,500 x g, 3 min, RT), and collect supernatant; the increase in volume allows for a more efficient removal of proteins duringvortexing.
- Concentrate sample back to 50 μl so SDS concentration is back to 1% (approximately 45 min at 45 °C using Speed-Vac). At this point, the samples can be mixed with loading buffer and run on an SDS PAGE gel. Proceed to step B1. If the samples are being prepared for massspectrometry analysis, the proteins can be reduced and alkylated prior to separation. Reduce the eluted proteins by adding DTT to a final concentration of 10 mM and boil for 2 min (for 50 μl, add 0.5 μl of 1 M DTT stock solution).
- Add iodoacetamide to a final concentration of 50 mM (for 50 μl, add 2.5 μl of 1 M IAA stocksolution) and incubate for 30 min at room temperature in the dark.
- Grow N. meningitidis from frozen stock: Obtain a small amount of frozen culture with a sterile 200 µl tip, inoculate Columbia agar base (CAB) plate and spread to give single colonies. Incubate overnight (~18 h) at 35 °C with 5% CO2. Collect 20-30 colonies and spread over the entire surface of a CAB plate, using a sterile loop, and incubate for 4 h at 35 °C with 5% CO2. Individual colonies will not be seen at this stage.
- 1D SDS-PAGE and sample digestion
- Mix reduced, alkylated samples with loading buffer (item 23 of Materials and Reagents list) andboil for 5 min.
- Separate proteins by SDS-PAGE (12%, 1 mm thick, 10 cm long).
- Stain with SimplyBlueTM for 1 h and de-stain with ultra-pure water (according to manufacturer’s instructions–see Figure 1).

Figure 1. Representative image of immunoprecipitated (IP) surface proteins of the meningococcus. Reproduced from Newcombe et al. (2014). - Cut gel lanes in as many slices as necessary based on sample protein concentration and place slices in low protein bind tubes.
Note: A full gel lane can be cut into 20 slices, however, it may only be necessary to run a shortened gel and cutting it into 5 slices. - Wash gel slices with 50% acetonitrile in 25 mM ammonium bicarbonate twice with vortexing for 15 min at room temperature to remove stain–use enough volume to cover the gel slices.
- Dehydrate gel slice by incubation with 100% acetonitrile (20 min, room temperature).
- Dry gel pieces using a Speed-Vac to dryness (about 20 min).
- Rehydrate in 25 mM ammonium bicarbonate containing modified trypsin (12.5 ng/μl–the final volume will depend on the size of the gel slice, 4 °C for 10 min). If required add more 25 mM ammonium bicarbonate to cover gel pieces, incubate overnight at 37 °C.
- Remove digest solution, extract digested proteins by incubation with 30 μl 0.1% formic acid/50% acetonitrile for 20-30 min with vortexing followed by 5 min in a sonicating bath (40 kHz)–repeat once and collect supernatant each time.
- Completely dry extracted digested proteins using Speed-Vac and resuspend in 20 μl water. Store at -20 °C.
- Mix reduced, alkylated samples with loading buffer (item 23 of Materials and Reagents list) andboil for 5 min.
- Liquid chromatography-electrospary ionization-MS/MS (please see Notes)
- Separate peptides using Ultimate 3000 HPLC and C18 pepman (Dionex) column at 40 °C, 350 nl/min, gradient of 2-50% solvent B for 30 min, 90% solvent B for 5 min–controlled by Chromeleon software.
- Set QSTAR XL to 2.3 kV electrospray, curtain gas at 8 arbitrary units, and 1 sec survey from 400 to 1,200 m/z with charge states 2-4.
- Acquire MS data with Information Data Acquisition with Analyst QS 1.1 software.
- Select most intense product ions with m/z of between 65 and 1,200 amu for MS/MS.
- Separate peptides using Ultimate 3000 HPLC and C18 pepman (Dionex) column at 40 °C, 350 nl/min, gradient of 2-50% solvent B for 30 min, 90% solvent B for 5 min–controlled by Chromeleon software.
- Database analysis (please see Notes)
- Submit peak lists of MS/MS spectra to Mascot Server (v2.2.04; Matrix Science) and analyse using the MS/MS Ions search programme.
- Allow up to one missing trypsin cleavage with fixed modification of carbamidomethyl (C) and variable modification of oxidation (M).
- Set peptide and MS/MS fragment tolerance to 1.2 and 0.6 Da respectively and select MH22+ and MH33+ as the precursor charge states.
- Search identified proteins against the National Center for Biotechnology Information nonredundant (NCBInr) database with the taxonomy set N. meningitidis (taxonomy code 487).Analyse the data with the appropriate database for the organism being used.
- Submit peak lists of MS/MS spectra to Mascot Server (v2.2.04; Matrix Science) and analyse using the MS/MS Ions search programme.
Data analysis
Once a list of identified proteins is obtained it should be checked to ensure all proteins are surface exposed. This can be done using cellular localization prediction tools such as PSORTb v3.0 (Yu etal., 2010), CELLO v2.5 (Yu et al., 2004 and 2006) or Gneg-mPLoc (Shen and Chou, 2010).Searching for signal peptides or lipidation can also be useful. We recommend the following tools:LipoP 1.0 (Juncker, 2003), SignalP 4.1 (Petersen et al., 2011), Signal-CF (Chou and Shen, 2007), and PrediSi (Hiller et al., 2004). To further assess protein location prediction, researchers are encouraged to look for β-barrel proteins and dismiss proteins containing transmembrane α-helix (beware that this conformational ‘rule’ does not apply to lipoproteins, only outer membrane proteins).
There are several techniques to assess experimentally the location of predicted proteins and they vary according to the bacterium being studied. Cell fractionation followed by Western blot, indirect immunofluorescence (unfixed samples), and cell surface proteolysis among other techniques may be suitable as well. To assess the localization of candidate proteins of the meningococcus, we produced serum against candidate proteins, probed intact bacteria with this serum, and added FITC-conjugated secondary antibodies and analysed single-cells by flow cytometry. The detection of fluorescence would confirm the surface exposure of a given candidate protein as demonstrated in Figure 2. Please refer to Newcombe et al., 2014 for more details on this technique and othertechniques we used.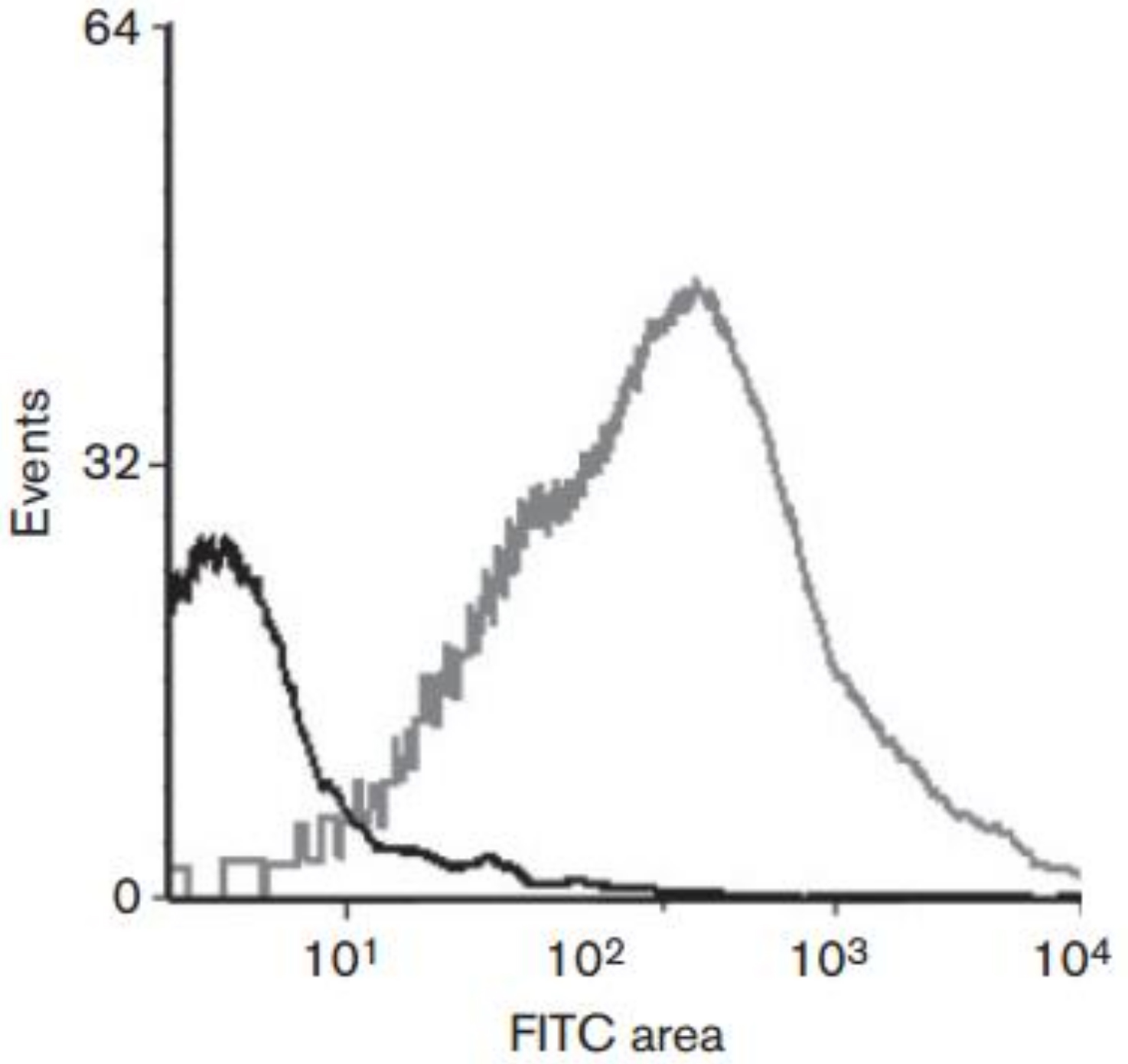
Figure 2. Representative data of flow cytometry experiment to assess whether a candidate protein is surface exposed or not. Cells probed with sera against surface exposed proteins and FITC show a stronger signal (grey line) than the negative control (black line). Reproduced from Newcombe et al. (2014).
Notes
- Serum samples should be selected based on immunity against the bacteria of interest, this canbe healthy sera which has shown to be confer immunity or sera specifically raised against the target organism. The antibody titre against the antigen will determine the amount of serum required to immunoprecipitate sufficient antigenic proteins.
- In addition to SDS-PAGE of immunoprecipitated proteins, we also recommend performing Western blots to confirm the presence of immunogenic proteins. The presence of other protein bands would indicate the extraction of non-specific proteins. It is essential to run controls including an antibody negative sample. It will be necessary to ensure the serum contains antibodies to the bacteria. The titre of the serum will affect the amount required to immunoprecipitate detectable levels of specific immunogenic proteins. The use of serum against non-surface proteins (e.g., periplasmic proteins, inner membrane proteins, etc.) is also advised to ensure there are no contamination of proteins that are not surface exposed.
- Step A7 of immunoprecipitation can be done using the spin columns or carefully pipetting off the supernatant.
- Mass spectrometry parameters are specific to the instrumentation available to the researchers. We report here specifications that we used, but researchers should refer to their own equipment instructions to find out how to better adjust their equipment. There are also other ways to analyse MS files (Procedure D). Other software and methodologies are available. We demonstrate here what software we incorporated in our analysis. It is recommended that the immunoprecipitated sample be split and or multiple samples analysed to enhance the reliability of the data.
- The preparation of peptides for the mass spectrometer analysis, the mass spectrometer data collection, and subsequent data analysis will depend on the facility performing the analysis. If this is in house or to be outsourced discuss the sample preparation before preparing the samples. All facilities have set procedures to ensure maximum output.
Recipes
- CAB medium
39 g Columbia agar base
Distilled water to 1 L
Autoclave (121 °C, 15 min)
Cool to 50 °C and add 6% sterile defibrinated blood - PBS plus 1% BSA
1 g BSA
PBS to 100 ml final volume, keep refrigerated - Solubilization buffer
3.0028 g Tris, 50 mM, pH 7.8
4.38 g NaCl, 150 mM
0.146 g EDTA (or dilute from 500 mM stock solution), 1 mM
1% (v/v) Triton X-100
1 g sodium deoxycholate, 0.2%
0.5 g SDS (or dilute from 10% stock solution–weight in fume hood), 0.1%
Distilled water (to a final volume of 500 ml) - 1% SDS
1 g SDS
100 ml LC/MS water
Weight in fume hood - 1 M dithiothreitol stock solution
1.542 g DTT
LC/MS water to 10 ml, keep at -20 °C
Weight in fume hood - 1 M Iodoacetamide stock solution
1.84 g IAA
Distilled LC/MS water to 10 ml - 25 mM ammonium bicarbonate
0.998 g ammonium bicarbonate
LC/MS water to 500 ml - Acetonitrile (50% v/v) in 25 mM ammonium bicarbonate
50 ml acetonitrile
50 ml ammonium bicarbonate (see above) - Acetonitrile (50% v/v)
50 ml acetonitrile
50 ml LC/MS water - Solvent A for HPLC
2 ml acetonitrile
0.1 ml formic acid
97.9 ml LC/MS water - Solvent B for HPLC
90 ml acetonitrile
0.1 ml formic acid
9.9 ml LC/MS water - 0.1% formic acid/50% acetonitrile solution
0.1 ml formic acid
50 ml acetonitrile
49.9 ml LC/MS water
Acknowledgments
The authors declare no conflicts of interest. This protocol has been previously described by us(Newcombe et al., 2014). Sanofi Pasteur funded this study. CNPq and CAPES funded the adaptation to other Gram-negative species.
References
- Chou, K. C. and Shen, H. B. (2007). Signal-CF: a subsite-coupled and window-fusing approach for predicting signal peptides. Biochem Biophys Res Commun 357(3): 633-640.
- Hiller, K., Grote, A., Scheer, M., Munch, R. and Jahn, D. (2004). PrediSi: prediction of signal peptides and their cleavage positions. Nucleic Acids Res 32(Web Server issue): W375-379.
- Juncker, A. S., Willenbrock, H., Von Heijne, G., Brunak, S., Nielsen, H. and Krogh, A. (2003). Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12(8): 1652-1662.
- Mendum, T. A., Newcombe, J., McNeilly, C. L. and McFadden, J. (2009). Towards the immunoproteome of Neisseria meningitidis. PLoS One 4(6): e5940.
- Newcombe, J., Mendum, T. A., Ren, C. P. and McFadden, J. (2014). Identification of the immunoproteome of the meningococcus by cell surface immunoprecipitation and MS. Microbiology 160(Pt 2): 429-438.
- Petersen, T. N., Brunak, S., von Heijne, G. and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8(10): 785-786.
- Shen, H. B. and Chou, K. C. (2010). Gneg-mPLoc: a top-down strategy to enhance the quality of predicting subcellular localization of Gram-negative bacterial proteins. J Theor Biol 264(2): 326-333.
- Vogel, U., Taha, M. K., Vazquez, J. A., Findlow, J., Claus, H., Stefanelli, P., Caugant, D. A., Kriz, P., Abad, R., Bambini, S., Carannante, A., Deghmane, A. E., Fazio, C., Frosch, M., Frosi, G., Gilchrist, S., Giuliani, M. M., Hong, E., Ledroit, M., Lovaglio, P. G., Lucidarme, J., Musilek, M., Muzzi, A., Oksnes, J., Rigat, F., Orlandi, L., Stella, M., Thompson, D., Pizza, M., Rappuoli, R., Serruto, D., Comanducci, M., Boccadifuoco, G., Donnelly, J. J., Medini, D. and Borrow, R. (2013). Predicted strain coverage of a meningococcal multicomponent vaccine (4CMenB) in Europe: a qualitative and quantitative assessment. Lancet Infect Dis 13(5): 416-425.
- Wheeler, J. X., Vipond, C. and Feavers, I. M. (2007). Exploring the proteome of meningococcal outer membrane vesicle vaccines. Proteomics Clin Appl 1(9): 1198-1210.
- Yu, C. S., Chen, Y. C., Lu, C. H. and Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins 64(3): 643-651.
- Yu, C. S., Lin, C. J. and Hwang, J. K. (2004). Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci 13(5): 1402-1406.
- Yu, N. Y., Wagner, J. R., Laird, M. R., Melli, G., Rey, S., Lo, R., Dao, P., Sahinalp, S. C., Ester, M., Foster, L. J. and Brinkman, F. S. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26(13): 1608-1615.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
da Cunha, C. E. P., Newcombe, J., Dellagostin, O. A. and McFadden, J. (2017). Immunoprecipitation of Cell Surface Proteins from Gram-negative Bacteria. Bio-protocol 7(9): e2250. DOI: 10.21769/BioProtoc.2250.
Category
Microbiology > Microbial biochemistry > Protein
Biochemistry > Protein > Immunodetection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









