- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Explant Methodology for Analyzing Neuroblast Migration
Published: Vol 7, Iss 9, May 5, 2017 DOI: 10.21769/BioProtoc.2249 Views: 10041
Reviewed by: Khyati Hitesh ShahAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Derivation and Culture of Enriched Phrenic-Like Motor Neurons From Human iPSCs
Louise Thiry [...] Stefano Stifani
Jul 5, 2025 2289 Views

Isolation and Imaging of Microvessels From Brain Tissue
Josephine K. Buff [...] Sophia M. Shi
Aug 5, 2025 2636 Views

Utilizing EdU to Track Leukocyte Recruitment to the Brain
Zoie K. Lipfert [...] David P. Sullivan
Dec 5, 2025 1564 Views
Abstract
The subventricular zone (SVZ) in the mammalian forebrain contains stem/progenitor cells that migrate through the rostral migratory stream (RMS) to the olfactory bulb throughout adulthood. SVZ-derived explant cultures provide a convenient method to assess factors regulating the intermediary stage of neural stem/progenitor cell migration. Here, we describe the isolation of SVZ-derived RMS explants from the neonatal mouse brain, and the conditions required to culture and evaluate their migration.
Keywords: NeuroblastsBackground
The adult mammalian forebrain contains a neurogenic niche that lies alongside the lateral ventricle in rodents and humans alike, and is aptly named the subventricular zone (SVZ). In rodents, the SVZ is a thin ‘wedge’ of cells, covering the entire wall of the lateral ventricle (Mirzadeh et al., 2010; Paez-Gonzalez et al., 2014; Dixon et al., 2016). Within the SVZ the slow-dividing astrocyte-like type B cells differentiate into rapidly dividing type C neural progenitor cells (also known as transit amplifying cells) that give rise to doublecortin-positive type A neuroblasts, although oligodendrocytes and astrocytes are also capable of being produced (Garcia-Verdugo et al., 1998; Tavazoie et al., 2008; Rikani et al., 2013). In rodents, there are an estimated 10,000 to 30,000 neuroblasts produced daily. These neuroblasts form chains as they migrate through the rostral migratory stream (RMS) to the olfactory bulb (Lois and Alvarez-Buylla, 1994; Sun et al., 2010). Ablation studies suggest it takes approximately 2 days for fast dividing type C neural progenitor cells to populate the SVZ, and an additional 2.5 days for neuroblasts to appear (Doetsch et al., 1999). A small percentage of these neuroblasts are capable of migrating ectopically out of the RMS into surrounding tissues in naïve mice; however, this phenomenon is drastically increased following brain injury (Dixon et al., 2016). The ability of neuroblasts to redirect their migratory routes towards damaged tissues has been shown to have beneficial effects on brain recovery (Li et al., 2010; Dixon et al., 2015), which can occur as early as 3 days post-injury (Ramaswamy et al., 2005; Dixon et al., 2016).
The self-renewal capacity of stem cells in culture was first identified in 1992 by Reynolds and Weiss (Reynolds and Weiss, 1992). The authors used fine dissection to harvest a small piece of the adult mouse striatum, before trypsinizing, dissociating and culturing. This original protocol, and subsequent variations, are now widely used to grow neurospheres or monolayer cultures to assess factors regulating stem cell survival, proliferation and/or differentiation into neurons (Theus et al., 2012). These culturing systems rely on the presence of growth factors (i.e., fibroblast and epidermal growth factors) to maintain proliferative states, whereas the withdrawal of these factors induces rapid differentiation into mature neurons. Unfortunately, these conditions limit the ability to analyze factors that regulate type A neuroblasts, a transient stage between the stem cell and neuron. To counteract this limitation; pieces of SVZ-derived tissue can be harvested and cultured as explants in a Matrigel containing laminin and collagen, which maintains the neural stem cells in their neuroblast state, allowing them to migrate (Ward and Rao, 2005; Dixon et al., 2016). Furthermore, neuroblast migration from cultured SVZ explants has similar characteristics to those observed in the RMS. Here, we describe an RMS explant methodology, modified from Ward and colleague (Leong et al., 2011), used to study chain migration of SVZ-derived neuroblasts.
Materials and Reagents
- 35 mm cell culture dishes with 4 internal wells, each with a diameter of 10 mm (Greiner Bio One International, catalog number: 627170 )
- 10 cm cell culture dishes (Corning, catalog number: 353003 )
- Pre-chilled (-20 °C) tissue culture pipette filter tips
- 10 µl tips (Corning, Axygen®, catalog number: TF-300-R-S )
- 200 µl tips (Corning, Axygen®, catalog number: TF-200-R-S )
- 1,000 µl tips (Corning, Axygen®, catalog number: TF-1000-R-S )
- 5 ml serological pipettes and pipette-boy (VWR, catalog number: 612-3702 )
- 1.5 ml Eppendorf microcentrifuge tubes (VWR, catalog number: 211-0007 )
- 3.2 ml disposable transfer pipette (Thermo Fisher Scientific, catalog number: BER202-1S )
- 1 ml insulin syringe with detachable needle (BD, catalog number: 329651 )
- Ice tray and ice as available
- Small biohazard bags as available
- Marker pen (e.g., Sharpie) for labelling cell culture plates
- 1-2 postnatal day old C57Bl/6 wildtype mice (Animal Resources Centre, Australia) or from local animal supplier
- Absolute ethanol (Sigma-Aldrich, catalog number: 24102 )
- Hanks balanced salt solution (HBSS) (Thermo Fisher Scientific, GibcoTM, catalog number: 14170112 )
- Growth factor reduced Matrigel (Corning, catalog number: 356230 )
- Neurobasal medium (Thermo Fisher Scientific, GibcoTM, catalog number: 21103049 )
- B27 supplement x50 (Thermo Fisher Scientific, GibcoTM, catalog number: 17504044 )
- 200 mM glutamine (Thermo Fisher Scientific, GibcoTM, catalog number: 25030081 )
- Penicillin/streptomycin (10,000 U/ml) (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- Recombinant murine fibroblast growth factor (FGF) basic 1 mg/ml (PeproTech, catalog number: 450-33 )
- Recombinant murine epidermal growth factor (EGF) 1 mg/ml (PeproTech, catalog number: 315-09 )
- Paraformaldehyde (Sigma-Aldrich, catalog number: 158127 )
- Sodium phosphate dibasic (Na2HPO4) (Chem Supply, catalog number: SA026 )
- Sodium dihydrogen phosphate monohydrate (NaH2PO4·H2O) (Chem Supply, catalog number: SO03310500 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888 )
- Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: S5881 )
- DAPI (Thermo Fisher Scientific, GibcoTM, catalog number: D1306 )
- Donkey serum (Sigma Aldrich, catalog number: D9663 )
- Triton X-100 (Sigma-Aldrich, catalog number: X100 )
- Goat anti-doublecortin (DCX) antibody (Santa Cruz Biotechnology, catalog number: sc-8066 or sc-271390 )
Note: The authors used sc-8066 , this antibody has been discontinued. A suggested alternative from Santa Cruz Biotechnology is sc-271390 . - Cy3-conjugated donkey anti-goat antibody (Jackson ImmunoResearch, catalog number: 705-165-147 )
- 80% ethanol solution (see Recipes)
- Complete neurobasal medium (see Recipes)
- Complete Matrigel (see Recipes)
- 0.1 M phosphate buffered saline (PBS) (see Recipes)
- 4% paraformaldehyde (PFA) (see Recipes)
- PBS containing DAPI (see Recipes)
- Blocking buffer (see Recipes)
- Primary antibody (see Recipes)
- Secondary antibody (see Recipes)
Equipment
- Pipettes
20 µl pipette (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4642050 )
200 µl pipette (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4642080 )
1,000 µl pipette (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 4642090 ) - Biosafety cabinet, any brand/model
- Bottles, sterile glass or plastic, any brand, for tissue culture medium storage
- Millipore StericupTM sterile vacuum filter units (EMD Millipore, catalog number: SCGPU01RE )
- Dissection microscope (Olympus, or similar) with lamp (or cold light source)
- Dissecting scissors, 12.5 cm long, straight (Coherent Scientific, catalog number: 15922 )
- Iris scissors 10 cm long, 30° angle, supercut (Coherent Scientific, catalog number: 500046 )
- Dressing forceps, 15.5 cm long (Coherent Scientific, catalog number: 500363 )
- 2 x Dumont forceps #3, 12 cm long, 0.08 x 0.04 mm tips (Coherent Scientific, catalog number: 500337 )
- Dissecting spatula, 140 mm long, 3 wide mm blade (World Precision Instruments, catalog number: 501772 )
- Scalpel handle No. 3 with scalpel blade No. 15 (Coherent Scientific , catalog numbers: 500236 and 500242 )
- Dumont forceps #5, 11 cm long, 0.06 x 0.01 mm tips (Coherent Scientific, catalog number: 14095 )
- Humidified tissue culture incubator (5% CO2, 37 °C), any brand/model
- Refrigerator (4 °C), any brand/model
- Freezer (-20 °C), any brand/model
- Inverted fluorescent microscope and digital camera (Olympus, model: IX81 or similar)
Software
- Axiovision software v4.1 (Zeiss, Thornwood, NY) or similar
- GraphPad Prism (v4.03)
Procedure
- Prepare working area
- Disinfect equipment using 80% ethanol (see Recipe 1), and place into a biosafety cabinet: dissection tools, 35 mm and 10 cm culture plates, media bottles, serological pipettes (5 ml) and pipette-boy, ice tray and ice, barrier pipette tips and relevant pipettes, and dissection microscope.
- Label all required dishes and tubes appropriately.
- Disinfect equipment using 80% ethanol (see Recipe 1), and place into a biosafety cabinet: dissection tools, 35 mm and 10 cm culture plates, media bottles, serological pipettes (5 ml) and pipette-boy, ice tray and ice, barrier pipette tips and relevant pipettes, and dissection microscope.
- Brain dissection
- Decapitate postnatal 1-2 day old C57Bl/6 mouse pups using dissecting scissors (along green dotted line) (Figure 1A). Dispose of body into biohazard bag.
- Using iris scissors, cut the skin on the scalp mid-sagittally (purple arrow) and peel laterally to expose the skull. Using dressing forceps and iris scissors, cut away the excess skin (Figure 1B).
- Using iris scissors, make the same cut through the skull, and two perpendicular cuts (blue arrows; Figure 1B). Using Dumont forceps #3 peel back the skull between these cuts to expose the underlying brain.
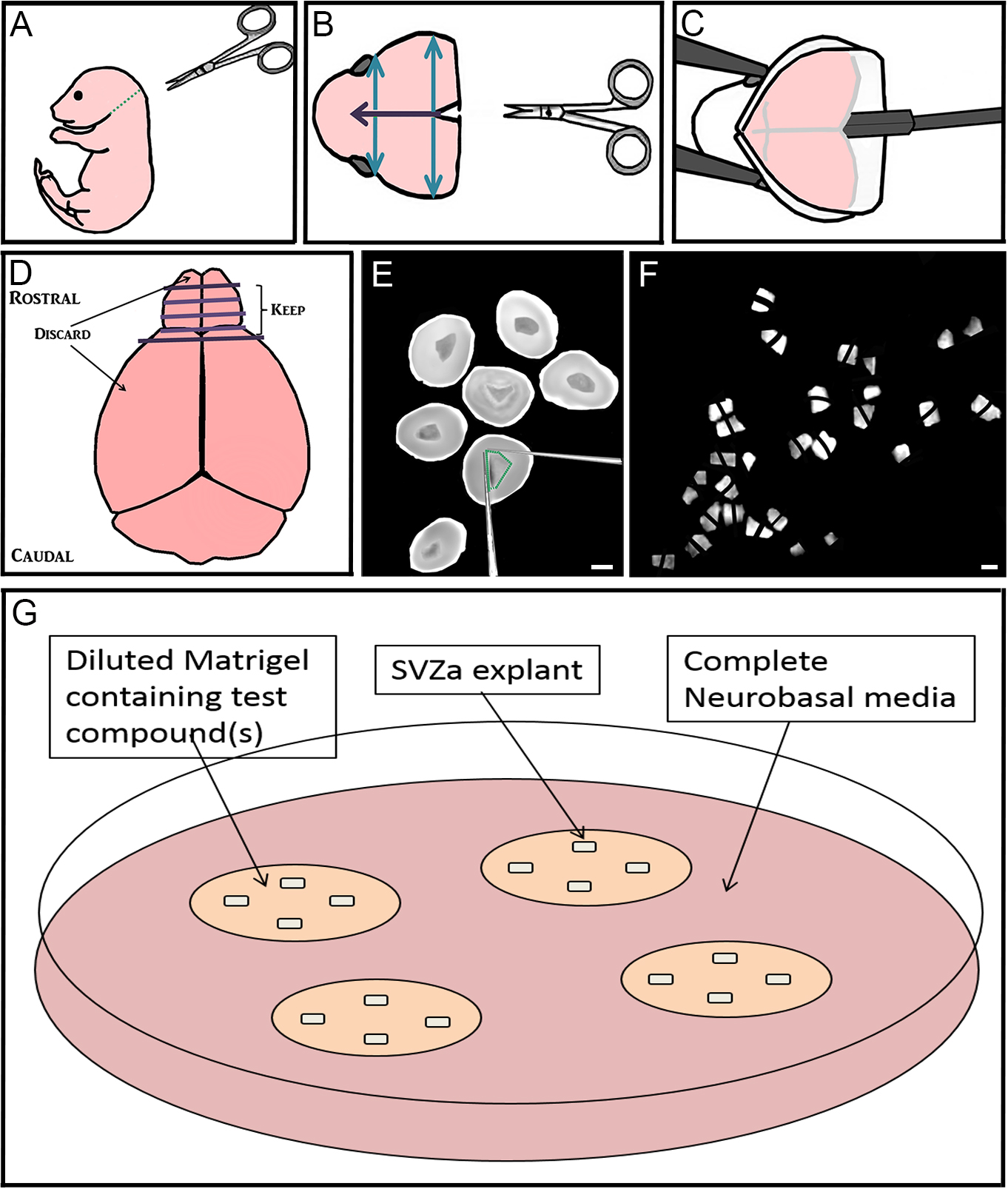
Figure 1. Step-by-step dissection procedure depicted in schematic and microscopic images. A-C. Removal of the brain from 1-2 day old mouse pups; D-F. Sectioning the rostral brain, dissection of RMS, and cutting RMS into many small pieces approximately 200-300 µm in diameter; G. Culturing up to 4 explants far apart in Matrigel with test compound(s) that are maintained in complete Neurobasal media. Bar represents 2 mm. - Using the flat part of the spatula, starting at the caudal end of the brain, gently lift the brain out of the skull (Figure 1C). Place the brain into a 10 cm culture dish containing 5 ml HBSS media. Dispose of mouse head into biohazard bag.
- Using two pairs of Dumont forceps #3, gently pull the meninges off the brain, starting with the olfactory bulbs.
- Decapitate postnatal 1-2 day old C57Bl/6 mouse pups using dissecting scissors (along green dotted line) (Figure 1A). Dispose of body into biohazard bag.
- Rostral migratory stream (RMS) dissection and explant production
- Using Dumont forceps #3 to hold the brain steady, use a scalpel blade at the rostral end of the brain to cut 3-4 coronal slices throughout the brain (Figure 1D). When using the scalpel blade, use a gentle back-and-forth sawing motion to preserve the morphology of the tissue.
- Using Dumont forceps #5, transfer rostral slices from both brain hemispheres containing RMS (which also contain migrating anterior subventricular zone [aSVZ] neuroblasts), into 35 mm dishes containing ice-cold HBSS (Figure 1E).
- To identify the RMS within these slices, angle the light source onto the center of the section so that the central RMS can be distinguished from the surrounding tissue. In the early postnatal mouse, the RMS will appear more translucent, dark and shiny than the surrounding tissue. The RMS should be located centrally, and at its widest point be about one third of the width of each coronal slice.
- Using two insulin syringe needles, dissect out the RMS by making cuts on the inside of its border (Figure 1E).
- Using the same syringe needles, cut the RMS into many small pieces approximately 200-300 µm wide, and set aside (Figure 1F).
Notes:
- The authors did not use a tissue/brain slicer to cut the brain into 3 or 4 coronal slices, although this equipment could assist others to cut more straight and even slices throughout the brain.
- To better visualize the RMS within a coronal slice, it may be important to re-arrange the light source so that the RMS appears darker from the surrounding tissue and thus can be distinguished more easily.
- For better experimental analysis, use the needles to cut the RMS into pieces that are approximately the same size and shape.
- One RMS may produce up to 50 explants.
- Using Dumont forceps #3 to hold the brain steady, use a scalpel blade at the rostral end of the brain to cut 3-4 coronal slices throughout the brain (Figure 1D). When using the scalpel blade, use a gentle back-and-forth sawing motion to preserve the morphology of the tissue.
- Explant experimentation
- Using ice-cold pipette tips and on top of ice tray, prepare complete Matrigel (see Recipe 3) by diluting Matrigel in complete neurobasal medium (see Recipe 2). To create different treatment conditions, add a maximum of 2 µl of the compound(s) to be tested into the diluted Matrigel (Figure 1G). Store on ice.
- Place a 4-well dish inside a 10 cm dish filled with ice to enable longer manipulation time of explants before the Matrigel polymerizes.
- Preparing one well at a time, using Dumont forceps #5, carefully pick up an explant and place into one well of the 4-well dish. Plate four RMS explants per well, and remove excess HBSS using a 10 µl pipette.
- Turn down the light source brightness to reduce any heat output (or use a cold light source illuminator).
- Using a scalpel blade, trim the tip of an ice-cold pipette tip to make a wider opening, and transfer 100 µl of diluted Matrigel onto explants in one well. Using the same tip, gently mix the Matrigel with the explants (without making any air bubbles), ensuring explants are entirely surrounded by the gel. Work quickly during this step otherwise the gel will begin to polymerize as it warms up under the heat of the microscope lamp.
- Using Dumont forceps #3, arrange the explants in the Matrigel so that they are far apart from each other, and far apart from the walls of the well.
- Repeat for other wells.
- Place two 35 mm 4-well dishes (containing explants and Matrigel) in a 10 cm dish and incubate (5% CO2, 37 °C) for 15 min in order for the Matrigel to polymerize.
- After polymerization, gently add 2 ml of complete neurobasal media to cover Matrigel and explants.
- Maintain cultures in a humidified incubator (5% CO2, 37 °C) for 72 h.
Notes:
- To prevent Matrigel from prematurely polymerizing, ensure manipulation is performed in ice-cold conditions using cell culture dishes and pipette tips pre-chilled at -20 °C.
- Mix the explants well in the Matrigel without making any bubbles, and position the explants in the wells far apart from each other, and away from the walls of the well to allow migration in all directions.
- Once the Matrigel and explants have been placed in the well it may become difficult to move the explants within the Matrigel. To assist with this process, prepare one well at a time. Between wells it may be necessary to turn off the light source (i.e., eliminate any heat sources) for 2-3 min and place a 10 cm Petri dish filled with ice on top of the dissecting microscope stage to cool it down quickly.
- Using a marker pen, keep track of the 4 explants in each well by labelling the bottom of the well 1, 2, 3 and 4.
- Using ice-cold pipette tips and on top of ice tray, prepare complete Matrigel (see Recipe 3) by diluting Matrigel in complete neurobasal medium (see Recipe 2). To create different treatment conditions, add a maximum of 2 µl of the compound(s) to be tested into the diluted Matrigel (Figure 1G). Store on ice.
- Explant immunohistochemistry
- Warm 4% PFA (see Recipe 5) to 37 °C
- Use a disposable transfer pipette to gently transfer solutions on and off the explants as follows.
- Remove media and add 2 ml warm PFA. Incubate for 1 h at room temperature.
- Remove PFA, and wash explants with PBS containing DAPI (see Recipe 6).
- Incubate explants in 2 ml blocking buffer (see Recipe 7) for 30 min at room temperature.
- Remove blocking buffer and incubate in 2 ml primary antibody solution (see Recipe 8) overnight at 4 °C.
- The next day gently wash the explants three times with PBS.
- Incubate explants in 2 ml fluorescent secondary antibody (see Recipe 9) for 48 h at 4 °C.
- Gently wash explants again three times with PBS.
- Remove media and add 2 ml warm PFA. Incubate for 1 h at room temperature.
- Gently remove blocking buffer from wells.
- Warm 4% PFA (see Recipe 5) to 37 °C
Data analysis
- Explant imaging and analysis
- Photograph explants on an inverted fluorescent microscope at 4x magnification.
- Categorize neuroblast migration away from RMS explant into 3 groups, as follows:
1 Explants that only give rise to compact chains 2 Explants that exhibit mixed cell outgrowth (individual and chains) 3 Explants that only exhibit cells migrating individually - Grade the extent of chain and cell outgrowth using a semi-quantitative scale, as follows (Figures 2A-2E):
0 = No outgrowth 1 = Less than 10 chains/cells 2 = Approximately 10-50 chains/cells 3 = Approximately 50-100 chains/cells 4 = Extensive growth (greater than 100 chains/cells) 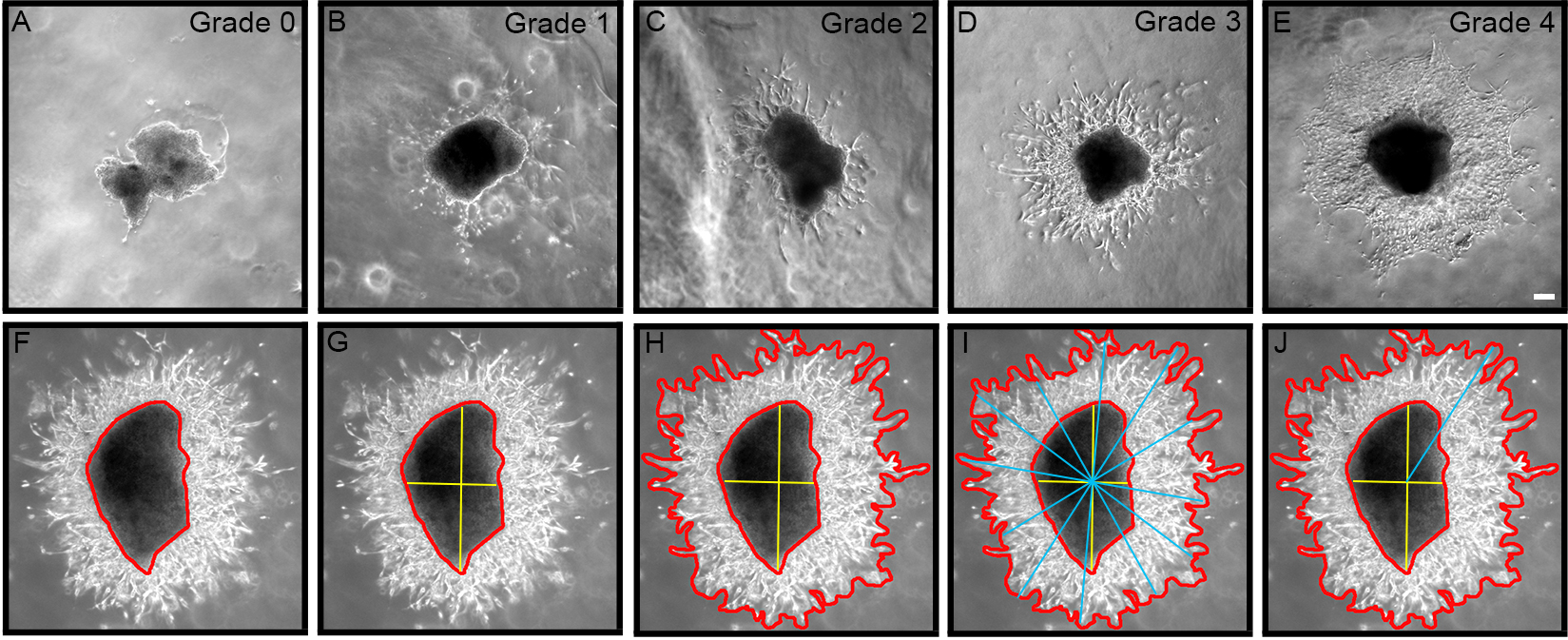
Figure 2. Grading chain/cell outgrowth from explant using semi-quantitative scale from 0-4. A. Zero (0) represents no growth; B. One represents less than 10 chains/cells; C. Two represents ~10-50 chains/cells; D. Three represents ~50-100 chains/cells; E. Four represents extensive growth > 100 chains/cells. F-J. Outgrowth area is calculated by subtracting explant area from total growth area, and longest migration length is determined using a line drawing tool through the explant epicenter. Bar represents 0.2 mm. - For each explant, measure the area of migration and longest migration length using Axiovision software v4.1 (Zeiss, Thornwood, NY), or similar Image Analysis software (Figures 2F-2J). As the explant sizes are not all the same, the area is measured for each explant by subtracting the area of the explant body from the total area of outgrowth (Figure 2H), and then normalize the area of outgrowth by expressing it as a ratio of outgrowth area:explant body area.
- Longest migration length can also be measured by using a line drawing tool as previously described (Cregg et al., 2010). First, the explant epicenter must be identify (yellow bars; Figure 2G), then a series of bars are drawn across the entire area of outgrowth intersecting the explant epicenter (blue bars; Figure 2I). The longest line from epicenter to outgrowth perimeter represents the longest migration length (Figure 2J).
- Average all results across different explant treatments, and then average results across individual experiments, and graph mean ± SEM. Assess data for homogeneity of variance using GraphPad Prism (v4.03). If a normal distribution is found, analyze data using one-way ANOVA, making individual comparisons with the Bonferroni post-hoc test. Set significance at P < 0.05.
- Perform analysis on at least 10-12 explants per treatment condition, and repeat experiment at least 3 times to create replicates.
- Photograph explants on an inverted fluorescent microscope at 4x magnification.
Recipes
- 80% ethanol solution
80 ml pure ethanol
20 ml H2O - Complete neurobasal medium
50 ml neurobasal medium
1 ml B27 (50x)
1 ml glutamine (4 mM)
500 µl penicillin/streptomycin (10,000 U/ml)
0.5 µl bFGF (1 mg/ml)
0.5 µl EGF (1 ng/ml)
Filter media using sterile vacuum filter units and store in the bottles, or in 50 ml tissue culture sterile conical tubes - Complete Matrigel
3 ml growth factor reduced Matrigel
1 ml complete neurobasal medium
In advance, thaw Matrigel stock solution overnight at 4 °C and make 0.5 ml aliquots using cold pipette tips and cold Eppendorf tubes, then store at -20 °C. On the day of the explant assay, thaw individual tubes on ice and dilute in complete neurobasal medium - 0.1 M phosphate buffered saline (PBS)
16 mM Na2HPO4
4 mM Na2HPO4·H2O
150 mM NaCl
Adjust to pH 7.4 - 4% paraformaldehyde (PFA)
4 g paraformaldehyde
Make up to 100 ml with PBS
Heat to a maximum of 60 °C
Add a few drops of NaOH until the solution turns clear
Adjust to pH 7.4 - PBS containing DAPI
10 ml PBS
1 µl DAPI - Blocking buffer
5 ml donkey serum (5%, v/v)
0.5 ml Triton X-100 (0.5%, v/v)
Make up to 100 ml with PBS - Primary antibody
2 µl Goat anti-doublecortin (DCX) antibody
1 ml blocking buffer - Secondary antibody
2 µl Cy3-conjugated donkey anti-goat
1 ml PBS
Acknowledgments
Studies were supported by NIH/NINDS (DJL: NS049545 and NS30291) and an NH&MRC fellowship (AMT: 628344). The work was adapted from Dixon et al. (2016).
References
- Cregg, J. M., Wiseman, S. L., Pietrzak-Goetze, M. N., Smith, M. R., Jaroch, D. B., Clupper, D. C. and Gilbert, R. J. (2010). A rapid, quantitiative method for assessing axonal extension on biomaterial platforms. Tissue Eng Part C Methods 16(2):167-172.
- Dixon, K. J., Mier, J., Gajavelli, S., Turbic, A., Bullock, R., Turnley, A. M. and Liebl, D. J. (2016). EphrinB3 restricts endogenous neural stem cell migration after traumatic brain injury. Stem Cell Res 17(3): 504-513.
- Dixon, K. J., Theus, M. H., Nelersa, C. M., Mier, J., Travieso, L. G., Yu, T. S., Kernie, S. G. and Liebl, D. J. (2015). Endogenous neural stem/progenitor cells stabilize the cortical microenvironment after traumatic brain injury. J Neurotrauma 32(11): 753-764.
- Doetsch, F., Garcia-Verdugo, J. M. and Alvarez-Buylla, A. (1999). Regeneration of a germinal layer in the adult mammalian brain. Proc Natl Acad Sci U S A 96(20): 11619-11624.
- Garcia-Verdugo, J. M., Doetsch, F., Wichterle, H., Lim, D. A. and Alvarez-Buylla, A. (1998). Architecture and cell types of the adult subventricular zone: in search of the stem cells. J Neurobiol 36(2): 234-248.
- Leong, S. Y., Faux, C. H., Turbic, A., Dixon, K. J. and Turnley, A. M. (2011). The Rho kinase pathway regulates mouse adult neural precursor cell migration. Stem Cells 29(2): 332-343.
- Li, B., Piao, C. S., Liu, X. Y., Guo, W. P., Xue, Y. Q., Duan, W. M., Gonzalez-Toledo, M. E. and Zhao, L. R. (2010). Brain self-protection: the role of endogenous neural progenitor cells in adult brain after cerebral cortical ischemia. Brain Res 1327: 91-102.
- Lois, C. and Alvarez-Buylla, A. (1994). Long-distance neuronal migration in the adult mammalian brain. Science 264(5162): 1145-1148.
- Mirzadeh, Z., Doetsch, F., Sawamoto, K., Wichterle, H. and Alvarez-Buylla, A. (2010). The subventricular zone en-face: wholemount staining and ependymal flow. J Vis Exp (39).
- Paez-Gonzalez, P., Asrican, B., Rodriguez, E. and Kuo, C. T. (2014). Identification of distinct ChAT(+) neurons and activity-dependent control of postnatal SVZ neurogenesis. Nat Neurosci 17(7): 934-942.
- Ramaswamy, S., Goings, G. E., Soderstrom, K. E., Szele, F. G. and Kozlowski, D. A. (2005). Cellular proliferation and migration following a controlled cortical impact in the mouse. Brain Res 1053(1-2): 38-53.
- Reynolds, B. A. and Weiss, S. (1992). Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255(5052): 1707-1710.
- Rikani, A. A., Choudhry, Z., Choudhry, A. M., Zenonos, G., Tariq, S. and Mobassarah, N. J. (2013). Spatially regulated adult neurogenesis. Ann Neurosci 20(2): 67-70.
- Sun, W., Kim, H. and Moon, Y. (2010). Control of neuronal migration through rostral migration stream in mice. Anat Cell Biol 43(4): 269-279.
- Tavazoie, M., Van der Veken, L., Silva-Vargas, V., Louissaint, M., Colonna, L., Zaidi, B., Garcia-Verdugo, J. M. and Doetsch, F. (2008). A specialized vascular niche for adult neural stem cells. Cell Stem Cell 3(3): 279-288.
- Theus, M. H., Ricard, J. and Liebl, D. J. (2012). Reproducible expansion and characterization of mouse neural stem/progenitor cells in adherent cultures derived from the adult subventricular zone. Curr Protoc Stem Cell Biol Chapter 2: Unit 2D 8.
- Ward, M. E. and Rao, Y. (2005). Investigations of neuronal migration in the central nervous system. Methods Mol Biol 294: 137-156.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Dixon, K. J., Turbic, A., Turnley, A. M. and Liebl, D. J. (2017). Explant Methodology for Analyzing Neuroblast Migration. Bio-protocol 7(9): e2249. DOI: 10.21769/BioProtoc.2249.
Category
Stem Cell > Adult stem cell > Neural stem cell
Cell Biology > Cell movement > Cell migration
Cell Biology > Tissue analysis > Tissue isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link